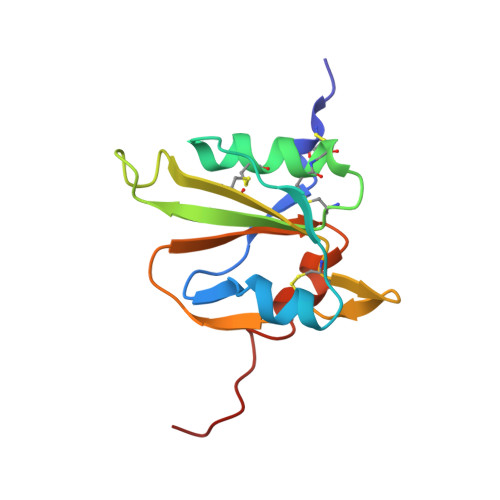
The solution structure of the MANEC-type domain from hepatocyte growth factor activator inhibitor-1 reveals an unexpected PAN/apple domain-type fold.
Hong, Z., Nowakowski, M., Spronk, C., Petersen, S.V., Andreasen, P.A., Kozminski, W., Mulder, F.A., Jensen, J.K.(2015) Biochem J 466: 299-309
- PubMed: 25510835
- DOI: https://doi.org/10.1042/BJ20141236
- Primary Citation of Related Structures:
2MSX - PubMed Abstract:
A decade ago, motif at N-terminus with eight-cysteines (MANEC) was defined as a new protein domain family. This domain is found exclusively at the N-terminus of >400 multi-domain type-1 transmembrane proteins from animals. Despite the large number of MANEC-containing proteins, only one has been characterized at the protein level: hepatocyte growth factor activator inhibitor-1 (HAI-1). HAI-1 is an essential protein, as knockout mice die in utero due to placental defects. HAI-1 is an inhibitor of matriptase, hepsin and hepatocyte growth factor (HGF) activator, all serine proteases with important roles in epithelial development, cell growth and homoeostasis. Dysregulation of these proteases has been causatively implicated in pathological conditions such as skin diseases and cancer. Detailed functional understanding of HAI-1 and other MANEC-containing proteins is hampered by the lack of structural information on MANEC. Although many MANEC sequences exist, sequence-based database searches fail to predict structural homology. In the present paper, we present the NMR solution structure of the MANEC domain from HAI-1, the first three-dimensional (3D) structure from the MANEC domain family. Unexpectedly, MANEC is a new subclass of the PAN/apple domain family, with its own unifying features, such as two additional disulfide bonds, two extended loop regions and additional α-helical elements. As shown for other PAN/apple domain-containing proteins, we propose a similar active role of the MANEC domain in intramolecular and intermolecular interactions. The structure provides a tool for the further elucidation of HAI-1 function as well as a reference for the study of other MANEC-containing proteins.
Organizational Affiliation:
*Department of Molecular Biology and Genetics, Danish-Chinese Centre for Proteases and Cancer, Aarhus University, 8000 Aarhus C, Denmark.