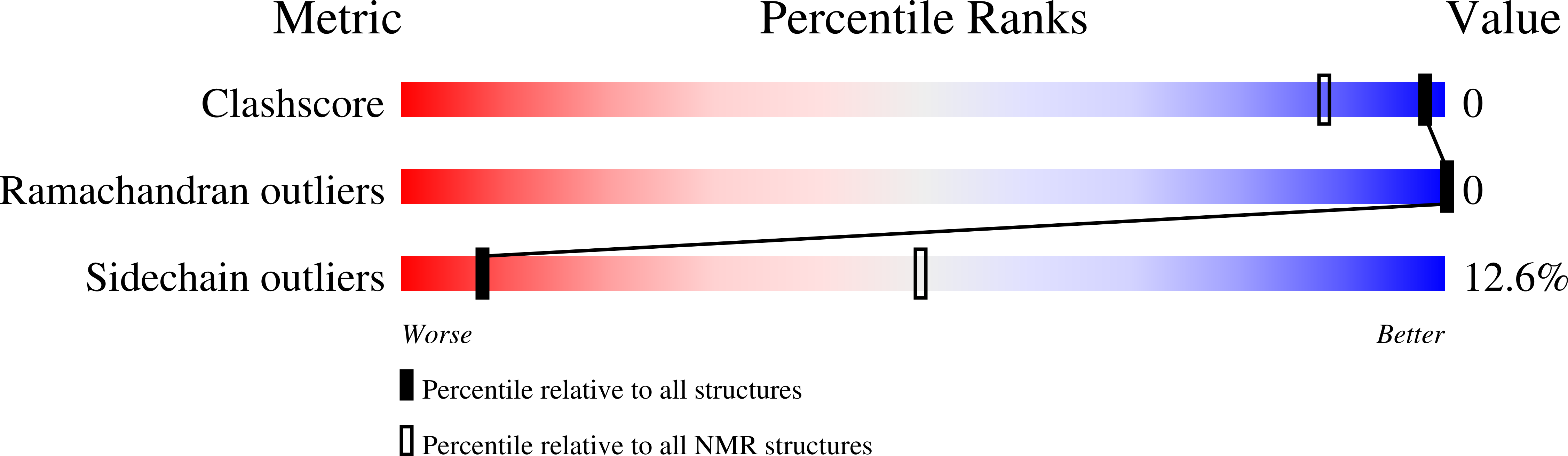Post-translational S-Nitrosylation Is an Endogenous Factor Fine Tuning the Properties of Human S100A1 Protein.
Lenarcic Zivkovic, M., Zareba-Koziol, M., Zhukova, L., Poznanski, J., Zhukov, I., Wyslouch-Cieszynska, A.(2012) J Biol Chem 287: 40457-40470
- PubMed: 22989881
- DOI: https://doi.org/10.1074/jbc.M112.418392
- Primary Citation of Related Structures:
2LLT, 2LLU - PubMed Abstract:
S100A1 protein is a proposed target of molecule-guided therapy for heart failure.
Organizational Affiliation:
National Institute of Chemistry, Slovenian NMR Centre, Hajdrihova 19, 1001 Ljubljana, Slovenia.