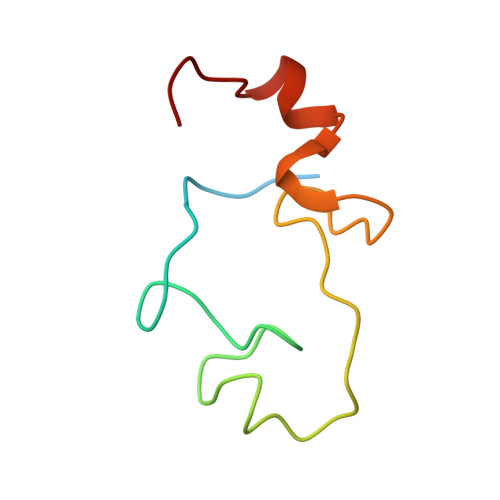
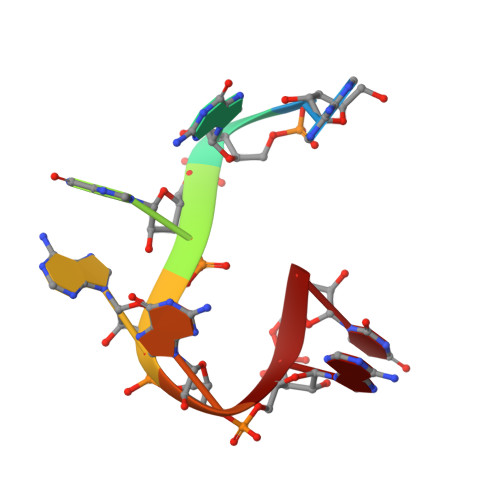
Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28.
Loughlin, F.E., Gebert, L.F., Towbin, H., Brunschweiger, A., Hall, J., Allain, F.H.(2011) Nat Struct Mol Biol 19: 84-89
- PubMed: 22157959
- DOI: https://doi.org/10.1038/nsmb.2202
- Primary Citation of Related Structures:
2LI8 - PubMed Abstract:
Lin28 inhibits the biogenesis of let-7 miRNAs through a direct interaction with the terminal loop of pre-let-7. This interaction requires the zinc-knuckle domains of Lin28. We show that the zinc knuckle domains of Lin28 are sufficient to provide binding selectivity for pre-let-7 miRNAs and present the NMR structure of human Lin28 zinc knuckles bound to the short sequence 5'-AGGAGAU-3'. The structure reveals that each zinc knuckle recognizes an AG dinucleotide separated by a single nucleotide spacer. This defines a new 5'-NGNNG-3' consensus motif that explains how Lin28 selectively recognizes pre-let-7 family members. Binding assays in cell lysates and functional assays in cultured cells demonstrate that the interactions observed in the solution structure also occur between the full-length protein and members of the pre-let-7 family. The consensus sequence explains several seemingly disparate previously published observations on the binding properties of Lin28.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zürich, Zürich, Switzerland.