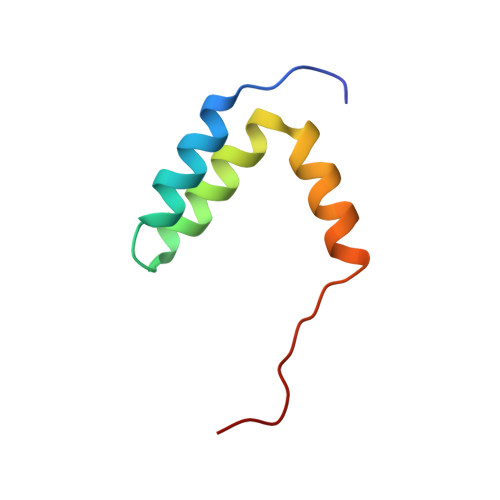
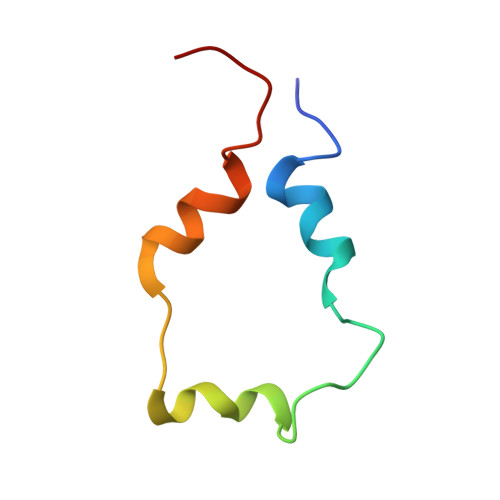
Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein.
Lee, C.W., Martinez-Yamout, M.A., Dyson, H.J., Wright, P.E.(2010) Biochemistry 49: 9964-9971
- PubMed: 20961098
- DOI: https://doi.org/10.1021/bi1012996
- Primary Citation of Related Structures:
2L14 - PubMed Abstract:
The activity and stability of the tumor suppressor p53 are regulated by interactions with key cellular proteins such as MDM2 and CBP/p300. The transactivation domain (TAD) of p53 contains two subdomains (AD1 and AD2) and interacts directly with the N-terminal domain of MDM2 and with several domains of CBP/p300. Here we report the NMR structure of the full-length p53 TAD in complex with the nuclear coactivator binding domain (NCBD) of CBP. Both the p53 TAD and NCBD are intrinsically disordered and fold synergistically upon binding, as evidenced by the observed increase in helicity and increased level of dispersion of the amide proton resonances. The p53 TAD folds to form a pair of helices (denoted Pα1 and Pα2), which extend from Phe19 to Leu25 and from Pro47 to Trp53, respectively. In the complex, the NCBD forms a bundle of three helices (Cα1, residues 2066-2075; Cα2, residues 2081-2092; and Cα3, residues 2095-2105) with a hydrophobic groove into which p53 helices Pα1 and Pα2 dock. The polypeptide chain between the p53 helices remains flexible and makes no detectable intermolecular contacts with the NCBD. Complex formation is driven largely by hydrophobic contacts that form a stable intermolecular hydrophobic core. A salt bridge between D49 of p53 and R2105 of NCBD may contribute to the binding specificity. The structure provides the first insights into simultaneous binding of the AD1 and AD2 motifs to a target protein.
Organizational Affiliation:
Department of Molecular Biology and The Skaggs Institute for Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, United States.