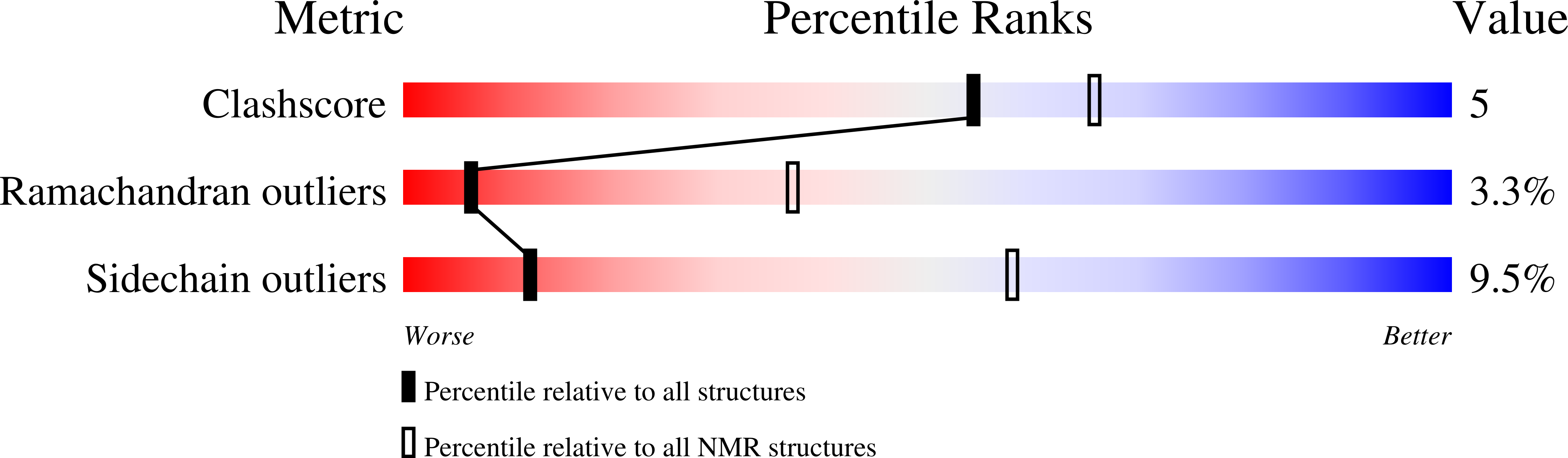Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase and its classification based on structural homologs
Motackova, V., Sanderova, H., Zidek, L., Novacek, J., Padrta, P., Svenkova, A., Korelusova, J., Jonak, J., Krasny, L., Sklenar, V.(2010) Proteins 78: 1807-1810
- PubMed: 20310067
- DOI: https://doi.org/10.1002/prot.22708
- Primary Citation of Related Structures:
2KRC
Organizational Affiliation:
National Centre for Biomolecular Research, Faculty of Science, Masaryk University, 611 37 Brno, Czech Republic.