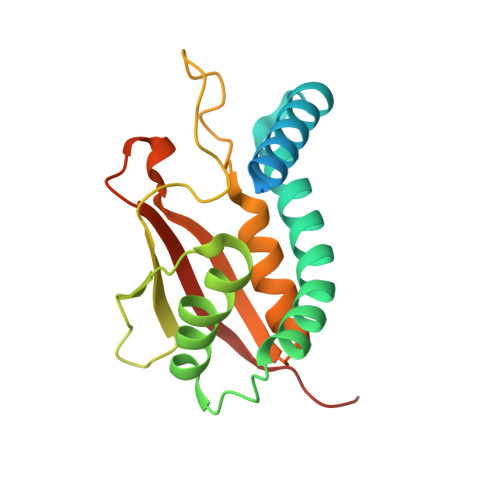
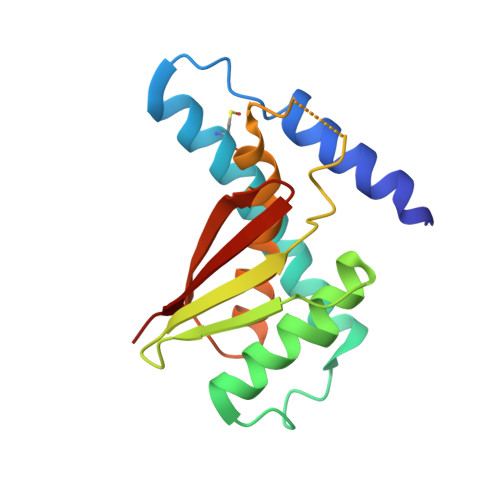
Structure of the Bet3-Tpc6B Core of Trapp: Two Tpc6 Paralogs Form Trimeric Complexes with Bet3 and Mum2.
Kummel, D., Muller, J.J., Roske, Y., Henke, N., Heinemann, U.(2006) J Mol Biol 361: 22
- PubMed: 16828797
- DOI: https://doi.org/10.1016/j.jmb.2006.06.012
- Primary Citation of Related Structures:
2CFH - PubMed Abstract:
The transport protein particle (TRAPP) complexes are involved in the tethering process at different trafficking steps of vesicle transport. We here present the crystal structure of a human Bet3-Tpc6B heterodimer, which represents a core sub-complex in the assembly of TRAPP. We describe a conserved patch of Tpc6 with uncharged pockets, forming a putative interaction interface for an anchoring moiety at the Golgi. The structural and functional comparison of the two paralogs Tpc6A and Tpc6B, only found in some organisms, indicates redundancy and added complexity of TRAPP architecture and function. Both iso-complexes, Bet3-Tpc6A and Bet3-Tpc6B, are able to recruit Mum2, a further TRAPP subunit, and we identify the alpha1-alpha2 loop regions as a binding site for Mum2. Our study reveals similar stability of the iso-complexes and similar expression patterns of the tpc6 variants in different mouse organs. These findings raise the possibility that the Tpc6 paralogs might contribute to the formation of two distinct TRAPP complexes that differ in function.
Organizational Affiliation:
Max-Delbrück Center for Molecular Medicine, Berlin, Germany.