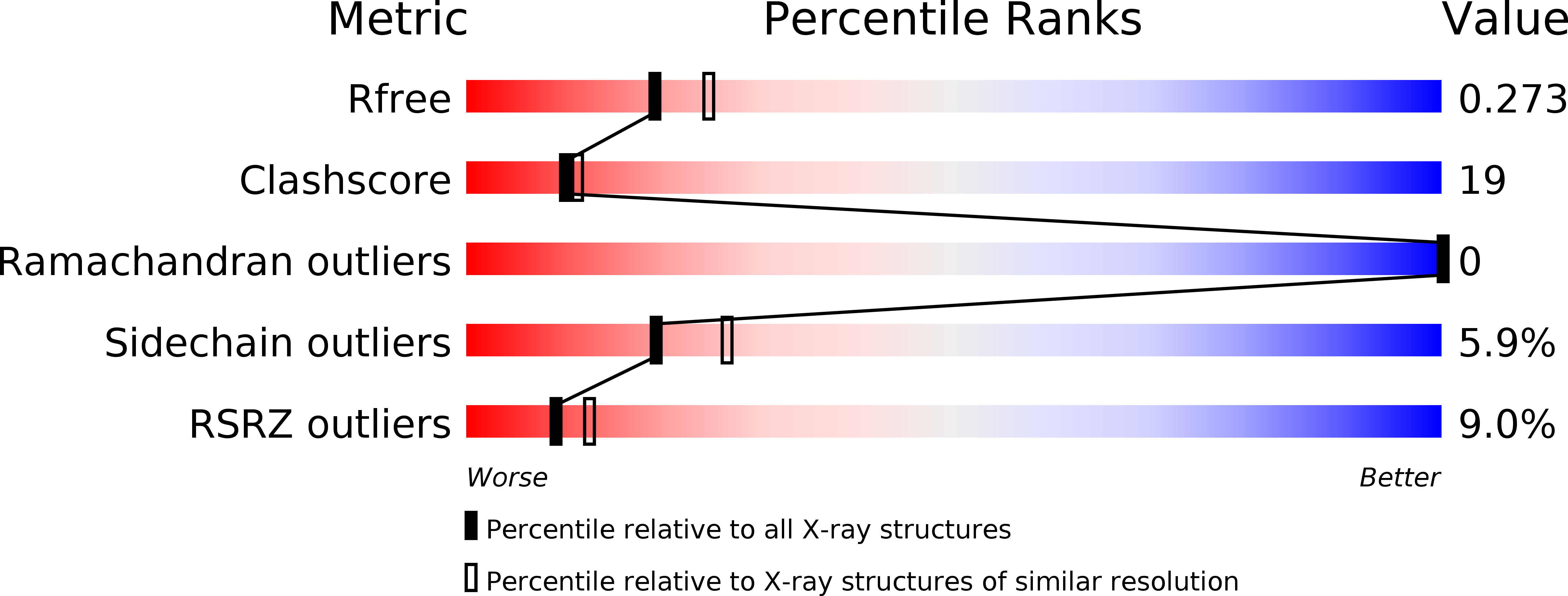
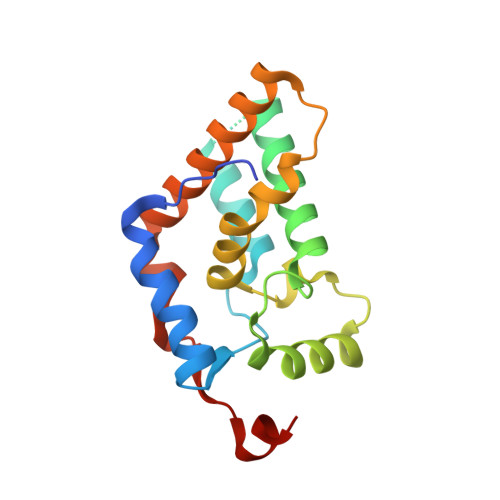
Multipurpose MRG domain involved in cell senescence and proliferation exhibits structural homology to a DNA-interacting domain.
Bowman, B.R., Moure, C.M., Kirtane, B.M., Welschhans, R.L., Tominaga, K., Pereira-Smith, O.M., Quiocho, F.A.(2006) Structure 14: 151-158
- PubMed: 16407074
- DOI: https://doi.org/10.1016/j.str.2005.08.019
- Primary Citation of Related Structures:
2AQL - PubMed Abstract:
The ubiquitous MRG/MORF family of proteins is involved in cell senescence, or the terminal loss of proliferative potential, a model for aging and tumor suppression at the cellular level. These proteins are defined by the approximately 20 kDa MRG domain that binds a plethora of transcriptional regulators and chromatin-remodeling factors, including the histone deacetylase transcriptional corepressor mSin3A and the novel nuclear protein PAM14, and they are also known components of the Tip60/NuA4 complex via interactions with the MRG binding protein (MRGBP). We present here the crystal structure of a prototypic MRG domain from human MRG15 whose core consists of two orthogonal helix hairpins. Despite the lack of sequence similarity, the core structure has surprisingly striking homology to a DNA-interacting domain of the tyrosine site-specific recombinases XerD, lambda integrase, and Cre. Site-directed mutagenesis studies based on the X-ray structure and bioinformatics identified key residues involved in the binding of PAM14 and MRGBP.
Organizational Affiliation:
Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, Texas 77030, USA.