Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
Martinez-Hackert, E., Anikeeva, N., Kalams, S.A., Walker, B.D., Hendrickson, W.A., Sykulev, Y.(2006) J Biol Chem 281: 20205-20212
- PubMed: 16702212
- DOI: https://doi.org/10.1074/jbc.M601934200
- Primary Citation of Related Structures:
1S8D, 1T1W, 1T1X, 1T1Y, 1T1Z, 1T20, 1T21, 1T22 - PubMed Abstract:
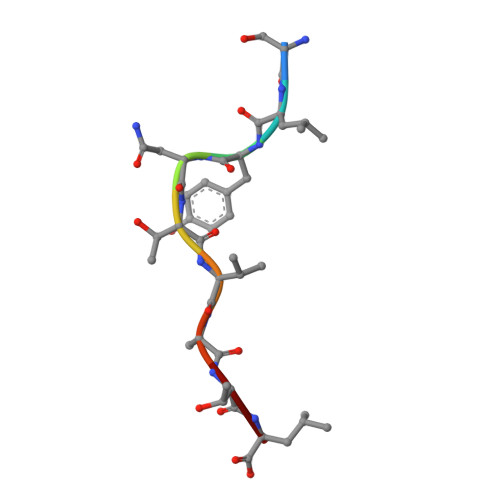
It is well established that even small changes in amino acid side chains of antigenic peptide bound to major histocompatibility complex (MHC) protein may completely abrogate recognition of the peptide-MHC (pMHC) complex by the T cell receptor (TCR). Often, however, several nonconservative substitutions in the peptide antigen are accommodated and do not impair its recognition by TCR. For example, a preponderance of natural sequence variants of the human immunodeficiency virus p17 Gag-derived peptide SLYNTVATL (SL9) are recognized by cytotoxic T lymphocytes, which implies that interactions with SL9 variants are degenerate both with respect to the class I MHC molecule and with respect to TCR. Here we study the molecular basis for this degenerate recognition of SL9 variants. We show that several SL9 variants bind comparably well to soluble HLA-A2 and to a particular soluble TCR and that these variants are active in the cognate cytotoxicity assay. Natural SL9 variation is restricted by its context in the HIV p17 matrix protein. High resolution crystal structures of seven selected SL9 variants bound to HLA-A2 all have remarkably similar peptide conformations and side-chain dispositions outside sites of substitution. This preservation of the peptide conformation despite epitope variations suggests a mechanism for the observed degeneracy in pMHC recognition by TCR and may contribute to the persistence of SL9-mediated immune responses in chronically infected individuals.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032, USA.