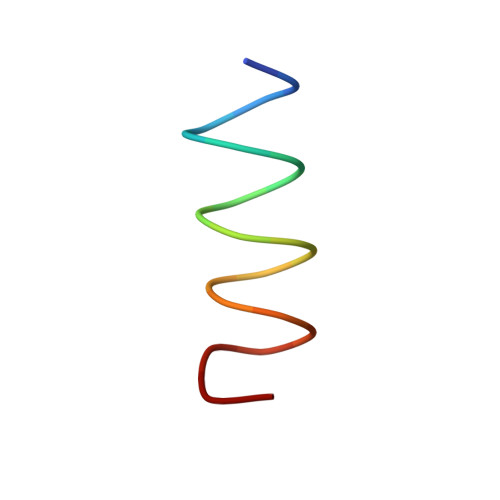
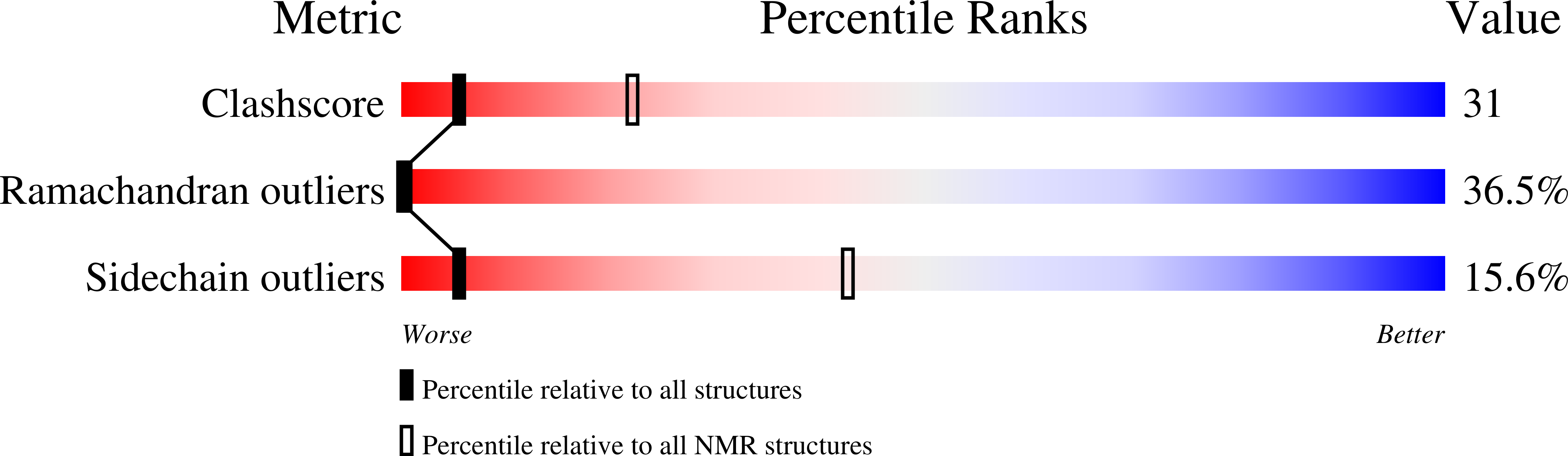Proline pipe helix: structure of the tus proline repeat determined by 1H NMR.
Butcher, D.J., Nedved, M.L., Neiss, T.G., Moe, G.R.(1996) Biochemistry 35: 698-703
- PubMed: 8547250
- DOI: https://doi.org/10.1021/bi952419l
- Primary Citation of Related Structures:
1SUT - PubMed Abstract:
The structure of a 22 amino acid peptide, TPPI [Nedved, M. L., Gottlieb, P. A., & Moe, G. R. (1994) Nucleic Acids Res. 22, 5024-5030], that is similar to the proline repeat segment of the replication arrest protein, Tus, has been determined by 1H NMR in 50% trifluroethanol. The structure is a novel left-handed helix having 5.56 residues per turn and a regular hydrogen bonding network that is limited to one side of the helix and contains a channel that runs down the helix axis. The latter feature gives the structure an overall pipe-like appearance; hence, the structure has been designated a proline pipe helix. The Tus proline pipe is also amphiphilic with one side consisting of proline and other nonpolar residues while the other side contains mostly basic and other polar residues. Tus and several other proteins that contain a similar proline repeat sequence are DNA binding proteins. It is shown here that the proline pipe helix of TPPI can be accommodated within the major grove of B-form DNA in a manner that positions nearly all of the basic residues near phosphate groups in the DNA backbone. The proline pipe helical motif may be a structural element of many other proteins including integral membrane receptor proteins.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Delaware, Newark 19716, USA.