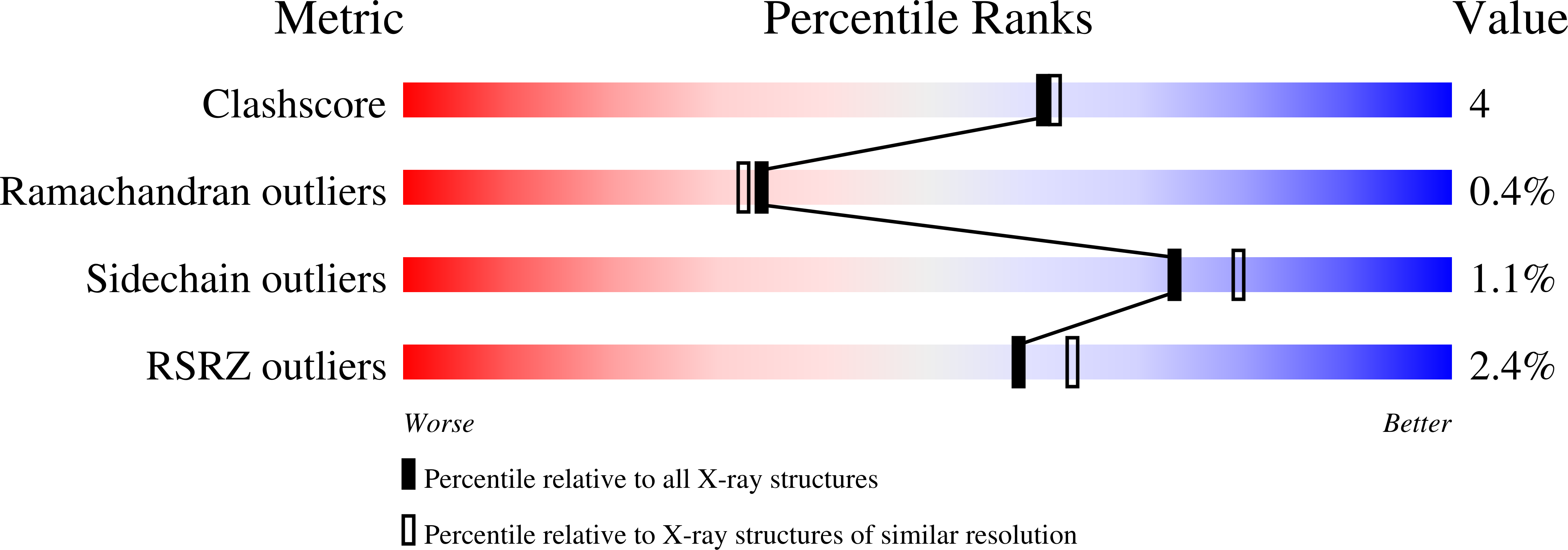
Mg2+ binding to the active site of EcoRV endonuclease: a crystallographic study of complexes with substrate and product DNA at 2 A resolution.
Kostrewa, D., Winkler, F.K.(1995) Biochemistry 34: 683-696
- PubMed: 7819264
- DOI: https://doi.org/10.1021/bi00002a036
- Primary Citation of Related Structures:
1RVA, 1RVB, 1RVC - PubMed Abstract:
The type II restriction endonuclease EcoRV was crystallized as a complex with the substrate DNA undecamer AAAGATATCTT (recognition sequence underlined). These crystals diffract to much better resolution (2 A) than was the case for the previously reported complex with the decamer GGGATATCCC [Winkler, F. K., Banner, D. W., Oefner, C., Tsernoglou, D., Brown, R. S., Heathman, S. P., Bryan, R. K., Martin, P. D., Petratos, K., & Wilson, K. S. (1993) EMBO J. 12, 1781-1795]. The crystal structure contains one dimer complex in the asymmetric unit and was solved by molecular replacement. The same kinked DNA conformation characteristic for enzyme-bound cognate DNA is observed. Crystals, soaked with Mg2+, show the essential cofactor bound at only one active site of the dimer, and the DNA is not cleaved. The Mg2+ has one oxygen from the scissile phosphodiester group and two carboxylate oxygens, one form Asp74 and one from Asp90, in its octahedral ligand sphere. The scissile phosphodiester group is pulled by 1 A toward the Mg2+. After substrate cleavage in solution, isomorphous crystals containing the enzyme--product--Mg2+ complex were obtained. In this structure, each of the 5'-phosphate groups is bound to two Mg2+. The kinked DNA conformation is essentially maintained, but the two central adenines, 3' to the cleavage sites, form an unusual cross-strand base stacking. The structures have been refined to R factors of 0.16 at 2.1-2.0 A resolution maintaining very good stereochemistry. On the basis of these structures and inspired by recent kinetic data [Vipond, I. B., & Halford, S. E. (1994) Biochemistry (second paper of three in this issue)], we have constructed a transition state model with two metals bound to the scissile phosphorane group.
Organizational Affiliation:
F. Hoffmann-LaRoche Ltd., Basel, Switzerland.