Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation.
Li, Y., Mitaxov, V., Waksman, G.(1999) Proc Natl Acad Sci U S A 96: 9491-9496
- PubMed: 10449720
- DOI: https://doi.org/10.1073/pnas.96.17.9491
- Primary Citation of Related Structures:
1QSS, 1QSY, 1QTM - PubMed Abstract:
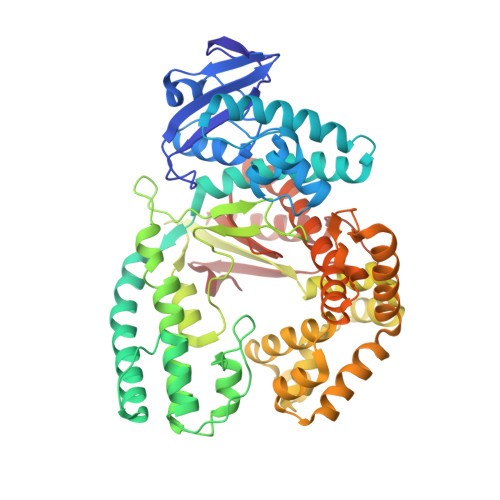
The Taq DNA polymerase is the most commonly used enzyme in DNA sequencing. However, all versions of Taq polymerase are deficient in two respects: (i) these enzymes incorporate each of the four dideoxynucleoside 5' triphosphates (ddNTPs) at widely different rates during sequencing (ddGTP, for example, is incorporated 10 times faster than the other three ddNTPs), and (ii) these enzymes show uneven band-intensity or peak-height patterns in radio-labeled or dye-labeled DNA sequence profiles, respectively. We have determined the crystal structures of all four ddNTP-trapped closed ternary complexes of the large fragment of the Taq DNA polymerase (Klentaq1). The ddGTP-trapped complex structure differs from the other three ternary complex structures by a large shift in the position of the side chain of residue 660 in the O helix, resulting in additional hydrogen bonds being formed between the guanidinium group of this residue and the base of ddGTP. When Arg-660 is mutated to Asp, Ser, Phe, Tyr, or Leu, the enzyme has a marked and selective reduction in ddGTP incorporation rate. As a result, the G track generated during DNA sequencing by these Taq polymerase variants does not terminate prematurely, and higher molecular-mass G bands are detected. Another property of these Taq polymerase variants is that the sequencing patterns produced by these enzymes are remarkably even in band-intensity and peak-height distribution, thus resulting in a significant improvement in the accuracy of DNA sequencing.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, 660 South Euclid Avenue, St. Louis, MO 63130, USA.