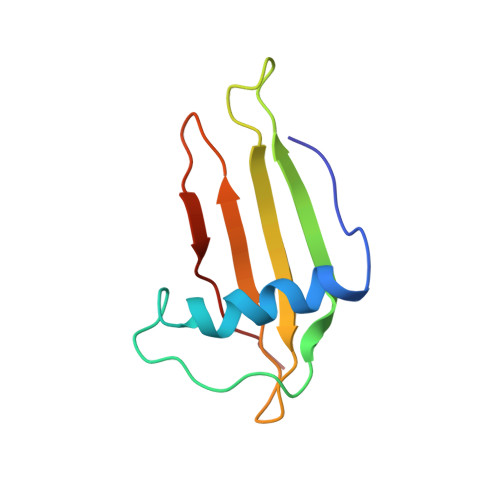
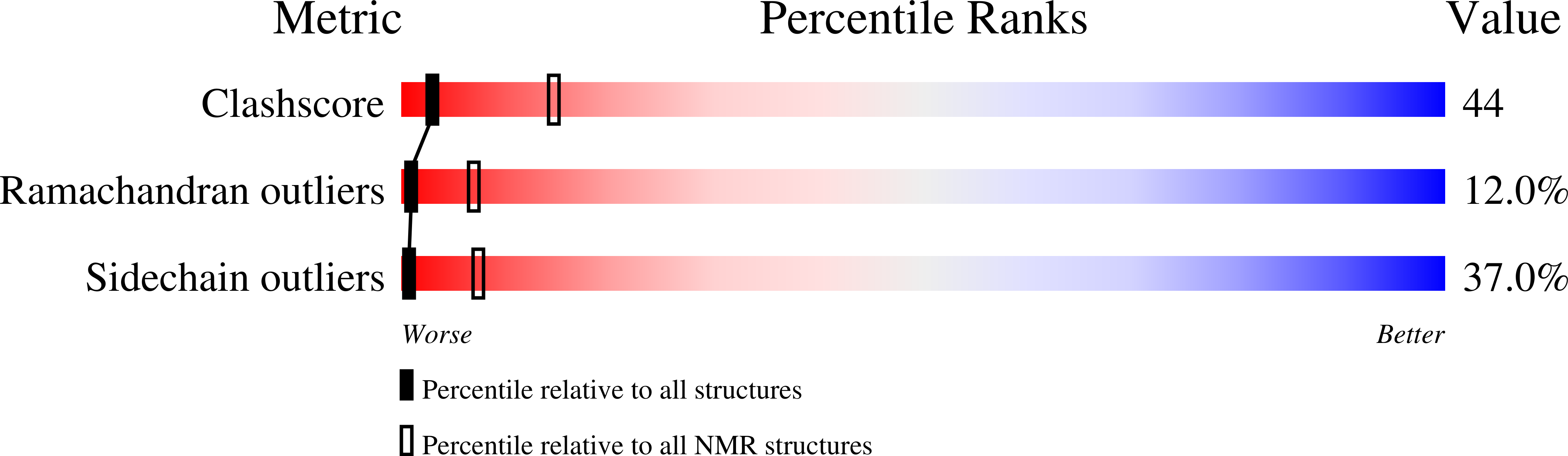Solution structure, backbone dynamics, and stability of a double mutant single-chain monellin. structural origin of sweetness.
Sung, Y.H., Shin, J., Chang, H.J., Cho, J.M., Lee, W.(2001) J Biol Chem 276: 19624-19630
- PubMed: 11279156
- DOI: https://doi.org/10.1074/jbc.M100930200
- Primary Citation of Related Structures:
1FUW - PubMed Abstract:
Single-chain monellin (SCM), which is an engineered 94-residue polypeptide, has been characterized as being as sweet as native two-chain monellin. Data from gel-filtration high performance liquid chromatography and NMR has proven that SCM exists as a monomer in aqueous solution. In order to determine the structural origin of the taste of sweetness, we engineered several mutant SCM proteins by mutating Glu(2), Asp(7), and Arg(39) residues, which are responsible for sweetness. In this study, we present the solution structure, backbone dynamics, and stability of mutant SCM proteins using circular dichroism, fluorescence, and NMR spectroscopy. Based on the NMR data, a stable alpha-helix and five-stranded antiparallel beta-sheet were identified for double mutant SCM. Strands beta1 and beta2 are connected by a small bulge, and the disruption of the first beta-strand were observed with SCM(DR) comprising residues of Ile(38)-Cys(41). The dynamical and folding characteristics from circular dichroism, fluorescence, and backbone dynamics studies revealed that both wild type and mutant proteins showed distinct dynamical as well as stability differences, suggesting the important role of mutated residues in the sweet taste of SCM. Our results will provide an insight into the structural origin of sweet taste as well as the mutational effect in the stability of the engineered sweet protein SCM.
Organizational Affiliation:
Department of Biochemistry and Protein Network Research Center, College of Science, Yonsei University, Seoul 120-740 Korea.