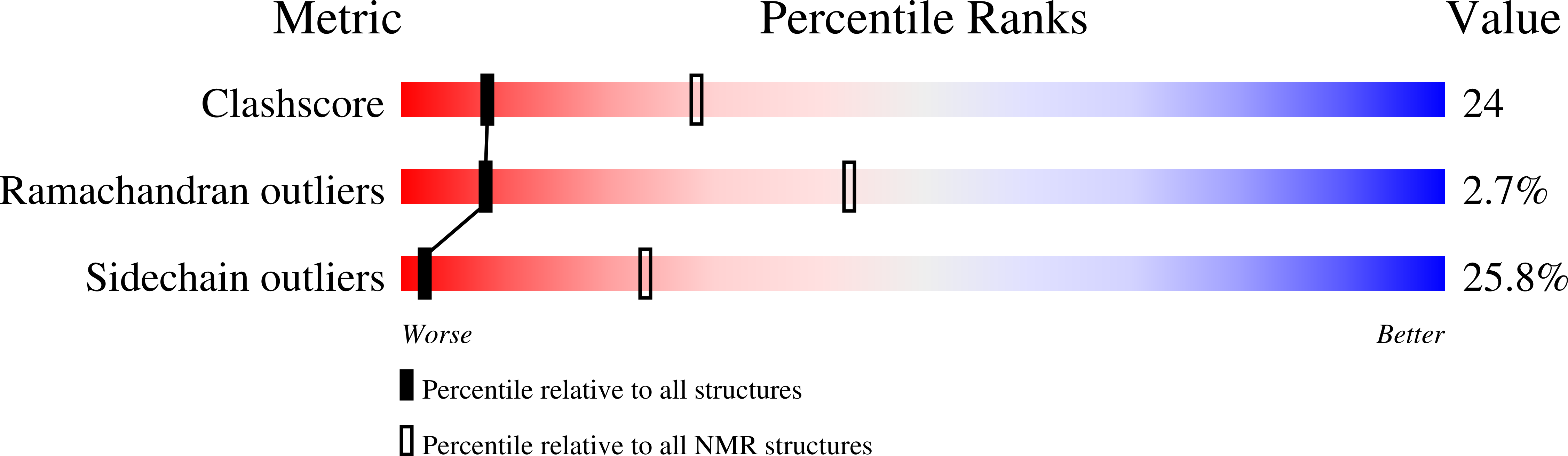Structural Basis for the Cytoskeletal Association of Bcr-Abl/c-Abl.
Hantschel, O., Wiesner, S., Guttler, T., Mackereth, C.D., Rix, L.L.R., Mikes, Z., Dehne, J., Gorlich, D., Sattler, M., Superti-Furga, G.(2005) Mol Cell 19: 461-473
- PubMed: 16109371
- DOI: https://doi.org/10.1016/j.molcel.2005.06.030
- Primary Citation of Related Structures:
1ZZP - PubMed Abstract:
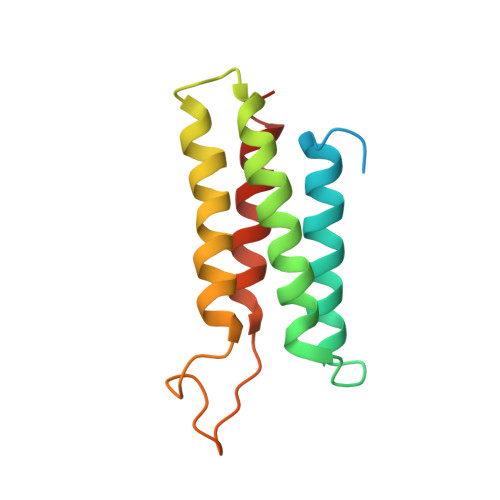
The Bcr-Abl tyrosine kinase causes different forms of leukemia in humans. Depending on its position within the cell, Bcr-Abl differentially affects cellular growth. However, no structural and molecular details for the anticipated localization determinants are available. We present the NMR structure of the F-actin binding domain (FABD) of Bcr-Abl and its cellular counterpart c-Abl. The FABD forms a compact left-handed four-helix bundle in solution. We show that the nuclear export signal (NES) previously reported in this region is part of the hydrophobic core and nonfunctional in the intact protein. In contrast, we could identify the critical residues of helix alphaIII that are responsible for F-actin binding and cytoskeletal association. We propose that these interactions represent a major determinant for both Bcr-Abl and c-Abl localization.
Organizational Affiliation:
Center for Molecular Medicine of the Austrian Academy of Sciences, Lazarettgasse 19/3, 1090 Vienna, Austria.