The Mechanism of Hsp90 Regulation by the Protein Kinase-Specific Cochaperone p50(Cdc37)
Roe, S.M., Ali, M.M.U., Meyer, P., Vaughan, C.K., Panaretou, B., Piper, P.W., Prodromou, C., Pearl, L.H.(2004) Cell 116: 87
- PubMed: 14718169
- DOI: https://doi.org/10.1016/s0092-8674(03)01027-4
- Primary Citation of Related Structures:
1US7 - PubMed Abstract:
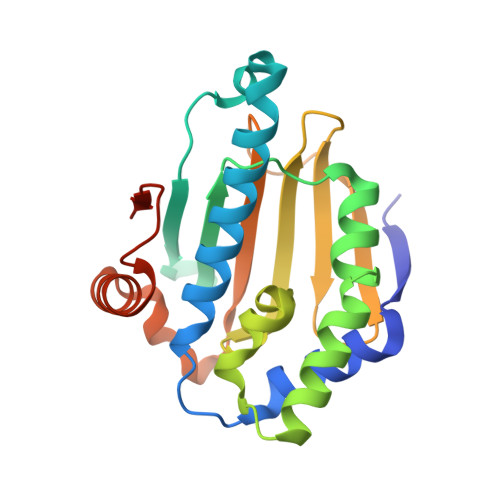
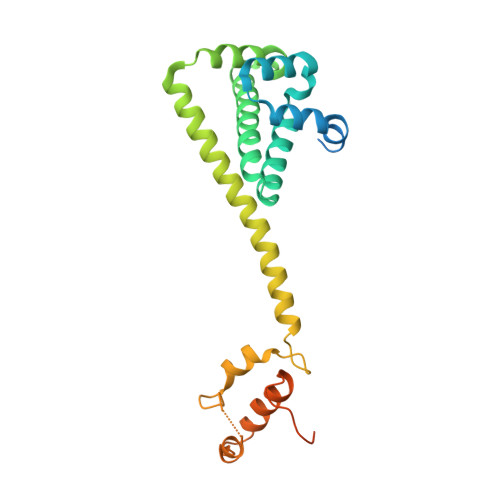
Recruitment of protein kinase clients to the Hsp90 chaperone involves the cochaperone p50(cdc37) acting as a scaffold, binding protein kinases via its N-terminal domain and Hsp90 via its C-terminal region. p50(cdc37) also has a regulatory activity, arresting Hsp90's ATPase cycle during client-protein loading. We have localized the binding site for p50(cdc37) to the N-terminal nucleotide binding domain of Hsp90 and determined the crystal structure of the Hsp90-p50(cdc37) core complex. Dimeric p50(cdc37) binds to surfaces of the Hsp90 N-domain implicated in ATP-dependent N-terminal dimerization and association with the middle segment of the chaperone. This interaction fixes the lid segment in an open conformation, inserts an arginine side chain into the ATP binding pocket to disable catalysis, and prevents trans-activating interaction of the N domains.
Organizational Affiliation:
Section of Structural Biology, The Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Road, London SW3 6JB, United Kingdom.