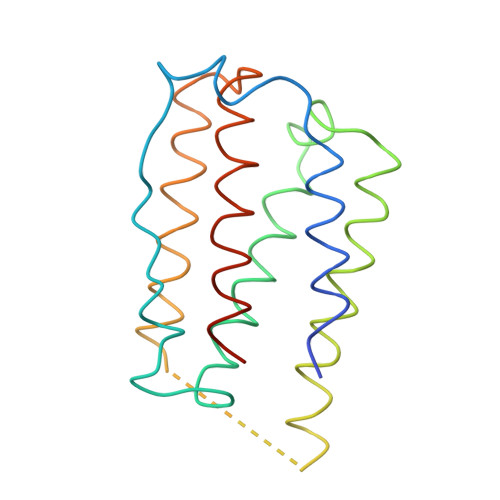
Zinc mediated dimer of human interferon-alpha 2b revealed by X-ray crystallography.
Radhakrishnan, R., Walter, L.J., Hruza, A., Reichert, P., Trotta, P.P., Nagabhushan, T.L., Walter, M.R.(1996) Structure 4: 1453-1463
- PubMed: 8994971
- DOI: https://doi.org/10.1016/s0969-2126(96)00152-9
- Primary Citation of Related Structures:
1RH2 - PubMed Abstract:
The human alpha-interferon (huIFN-alpha) family displays broad spectrum antiviral, antiproliferative and immunomodulatory activities on a variety of cell types. The diverse biological activities of the IFN-alpha's are conveyed to cells through specific interactions with cell-surface receptors. Despite considerable effort, no crystal structure of a member of this family has yet been reported, because the quality of the protein crystals have been unsuitable for crystallographic studies. Until now, structural models of the IFN-alpha's have been based on the structure of murine IFN-beta (muIFN-beta). These models are likely to be inaccurate, as the amino acid sequence of muIFN-beta differs significantly from the IFN-alpha's at proposed receptor-binding sites. Structural information on a huIFN-alpha subtype would provide an improved basis for modeling the structures of the entire IFN-alpha family.
Organizational Affiliation:
Center for Macromolecular Crystallography, University of Alabama at Brimingham 35294, USA.