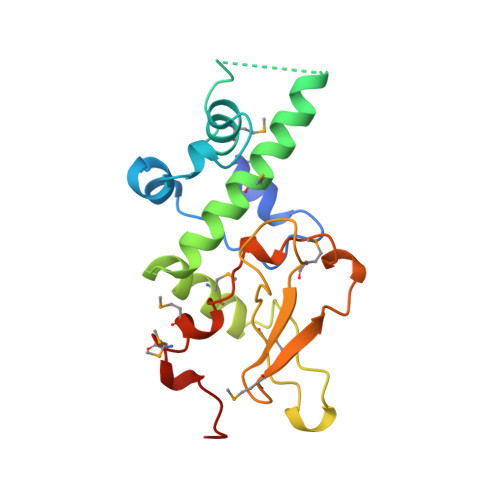
Crystal structure of the human protein kinase CK2 regulatory subunit reveals its zinc finger-mediated dimerization.
Chantalat, L., Leroy, D., Filhol, O., Nueda, A., Benitez, M.J., Chambaz, E.M., Cochet, C., Dideberg, O.(1999) EMBO J 18: 2930-2940
- PubMed: 10357806
- DOI: https://doi.org/10.1093/emboj/18.11.2930
- Primary Citation of Related Structures:
1QF8 - PubMed Abstract:
Protein kinase CK2 is a tetramer composed of two alpha catalytic subunits and two beta regulatory subunits. The structure of a C-terminal truncated form of the human beta subunit has been determined by X-ray crystallography to 1.7 A resolution. One dimer is observed in the asymmetric unit of the crystal. The most striking feature of the structure is the presence of a zinc finger mediating the dimerization. The monomer structure consists of two domains, one entirely alpha-helical and one including the zinc finger. The dimer has a crescent shape holding a highly acidic region at both ends. We propose that this acidic region is involved in the interactions with the polyamines and/or catalytic subunits. Interestingly, conserved amino acid residues among beta subunit sequences are clustered along one linear ridge that wraps around the entire dimer. This feature suggests that protein partners may interact with the dimer through a stretch of residues in an extended conformation.
Organizational Affiliation:
Laboratoire de Cristallographie Macromoléculaire, Institut de Biologie Structurale Jean-Pierre Ebel, CNRS/CEA, 41, rue Jules Horowitz, 38027 Grenoble Cedex 1, France.