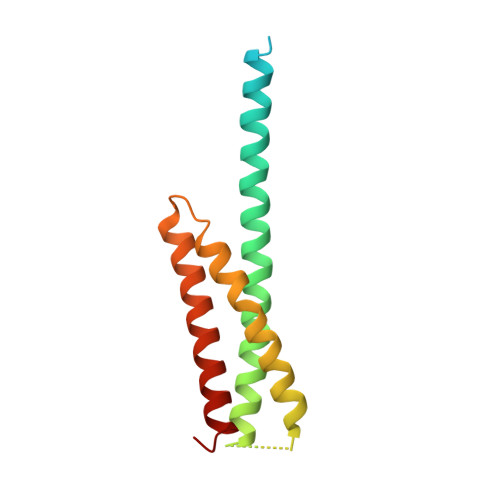
Structure of the GAT domain of human GGA1: a syntaxin amino-terminal domain fold in an endosomal trafficking adaptor.
Suer, S., Misra, S., Saidi, L.F., Hurley, J.H.(2003) Proc Natl Acad Sci U S A 100: 4451-4456
- PubMed: 12668765
- DOI: https://doi.org/10.1073/pnas.0831133100
- Primary Citation of Related Structures:
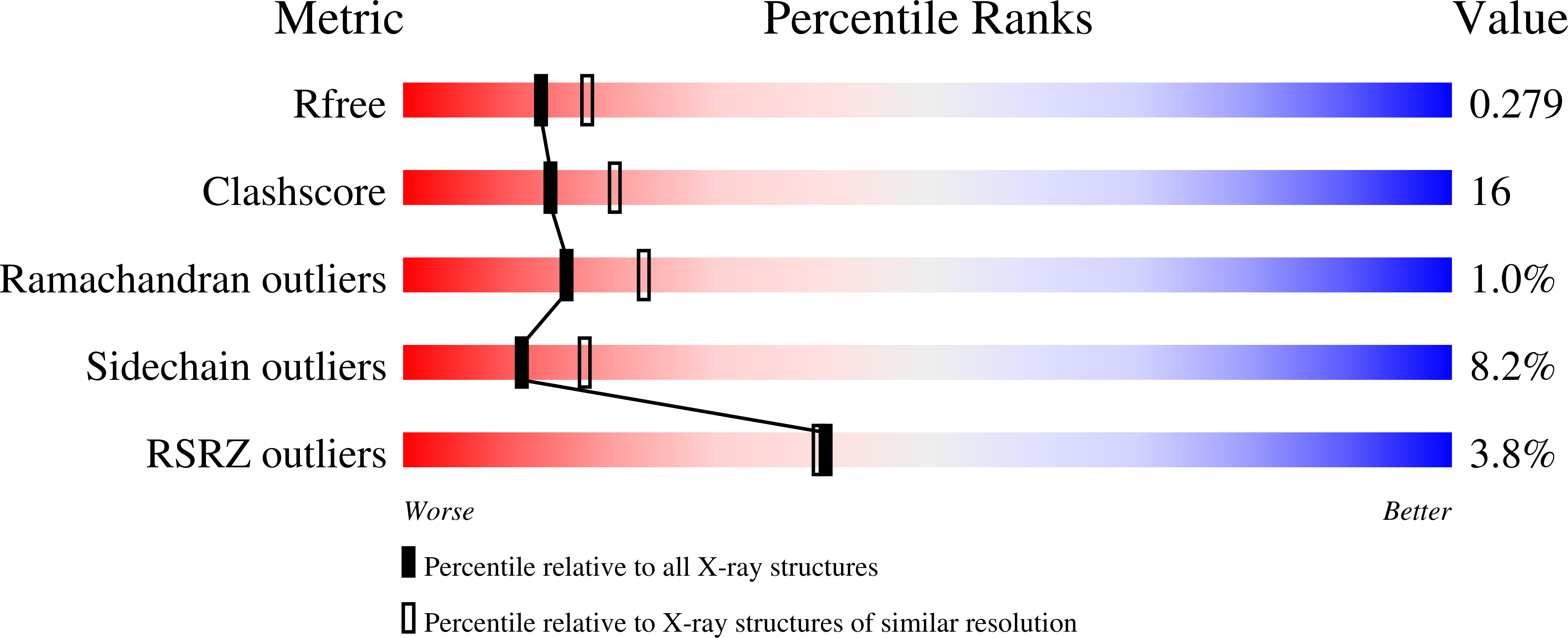
1NWM - PubMed Abstract:
The Golgi-associated, gamma-adaptin homologous, ADP-ribosylation factor (ARF)-interacting proteins (GGAs) are adaptors that sort receptors from the trans-Golgi network into the endosomallysosomal pathway. The GGAs and TOM1 (GAT) domains of the GGAs are responsible for their ARF-dependent localization. The 2.4-A crystal structure of the GAT domain of human GGA1 reveals a three-helix bundle, with a long N-terminal helical extension that is not conserved in GAT domains that do not bind ARF. The ARF binding site is located in the N-terminal extension and is separate from the core three-helix bundle. An unanticipated structural similarity to the N-terminal domain of syntaxin 1a was discovered, comprising the entire three-helix bundle. A conserved binding site on helices 2 and 3 of the GAT domain three-helix bundle is predicted to interact with coiled-coil-containing proteins. We propose that the GAT domain is descended from the same ancestor as the syntaxin 1a N-terminal domain, and that both protein families share a common function in binding coiled-coil domain proteins.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Department of Health and Human Services, Bethesda, MD 20892, USA.