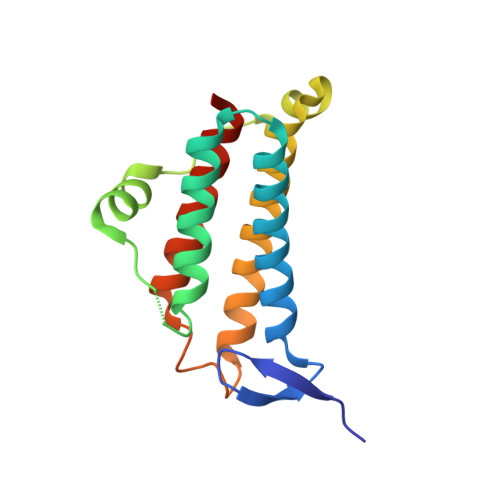
Crystal structure of the hypothetical protein TA1238 from Thermoplasma acidophilum: a new type of helical super-bundle.
Sanishvili, R., Pennycooke, M., Gu, J., Xu, X., Joachimiak, A., Edwards, A.M., Christendat, D.(2004) J Struct Funct Genomics 5: 231-240
- PubMed: 15704011
- DOI: https://doi.org/10.1007/s10969-005-3789-1
- Primary Citation of Related Structures:
1NIG - PubMed Abstract:
The crystal structure of the hypothetical protein TA1238 from Thermoplasma acidophilum was solved with multiple-wavelength anomalous diffraction and refined at 2.0 A resolution. The molecule consists of a typical four-helix antiparallel bundle with overhand connection. However, its oligomerization into a trimer leads to a coiled "super-helix" which is novel for such bundles. Its central feature, a six-stranded coiled coil, is also novel for proteins. TA1238 does not have strong sequence homologues in databases, but shows strong structural similarity with some proteins in the Protein Data Bank. The function could not be inferred from the sequence but the structure, with some rearrangement, bears some resemblance to the active site region of cobalamin adenosyltransferase (TA1434). Specifically, TA1238 retains Arg104, which is structurally equivalent to functionally critical Arg119 of TA1434. For such conformational change, the overhand connection of TA1238 might need to be involved in a gating mechanism that might be modulated by ligands and/or by interactions with the physiological partners. This allowed us to hypothesize that TA1238 could be involved in cobalamin biosyntheses.
Organizational Affiliation:
Biosciences, Argonne National Laboratory, Structural Biology Center, Midwest Center for Structural Genomics, 9700 South Cass Avenue, Argonne, IL 60439, USA.