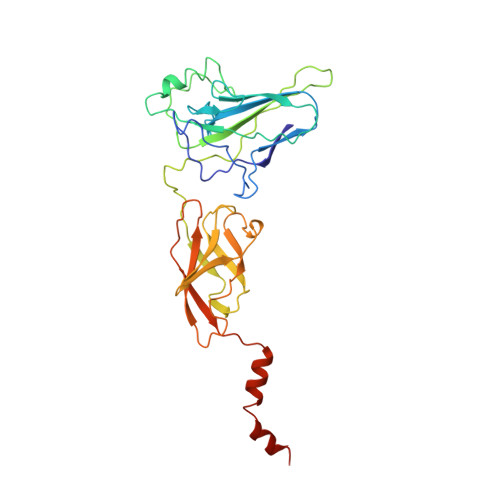
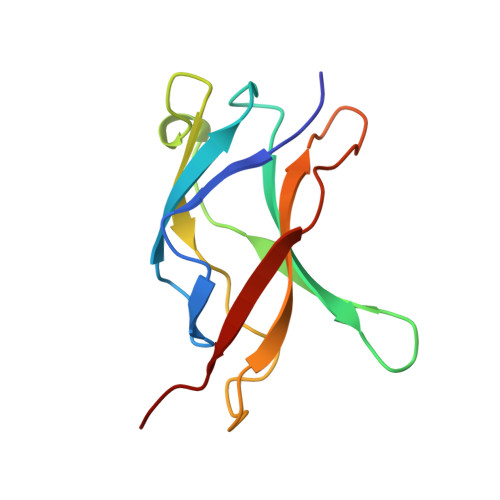
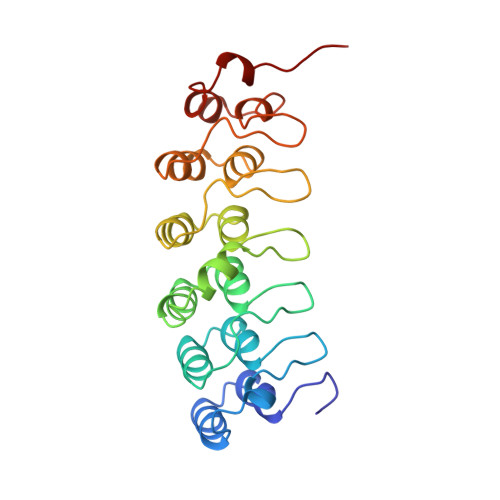
Structure of an IkappaBalpha/NF-kappaB complex.
Jacobs, M.D., Harrison, S.C.(1998) Cell 95: 749-758
- PubMed: 9865693
- DOI: https://doi.org/10.1016/s0092-8674(00)81698-0
- Primary Citation of Related Structures:
1NFI - PubMed Abstract:
The inhibitory protein, IkappaBalpha, sequesters the transcription factor, NF-kappaB, as an inactive complex in the cytoplasm. The structure of the IkappaBalpha ankyrin repeat domain, bound to a partially truncated NF-kappaB heterodimer (p50/ p65), has been determined by X-ray crystallography at 2.7 A resolution. It shows a stack of six IkappaBalpha ankyrin repeats facing the C-terminal domains of the NF-kappaB Rel homology regions. Contacts occur in discontinuous patches, suggesting a combinatorial quality for ankyrin repeat specificity. The first two repeats cover an alpha helically ordered segment containing the p65 nuclear localization signal. The position of the sixth ankyrin repeat shows that full-length IkappaBalpha will occlude the NF-kappaB DNA-binding cleft. The orientation of IkappaBalpha in the complex places its N- and C-terminal regions in appropriate locations for their known regulatory functions.
Organizational Affiliation:
Department of Molecular and Cellular Biology, Harvard University, Cambridge, Massachusetts 02138, USA.