Structural and Functional Analysis of the Actin Binding Domain of Plectin Suggests Alternative Mechanisms for Binding to F-Actin and Integrin Beta4
Garcia-Alvarez, B., Bobkov, A., Sonnenberg, A., de Pereda, J.M.(2003) Structure 11: 615-625
- PubMed: 12791251
- DOI: https://doi.org/10.1016/s0969-2126(03)00090-x
- Primary Citation of Related Structures:
1MB8 - PubMed Abstract:
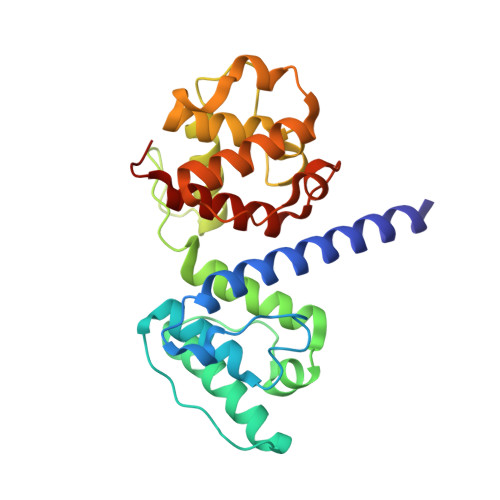
Plectin is a widely expressed cytoskeletal linker. Here we report the crystal structure of the actin binding domain of plectin and show that this region is sufficient for interaction with F-actin or the cytoplasmic region of integrin alpha6beta4. The structure is formed by two calponin homology domains arranged in a closed conformation. We show that binding to F-actin induces a conformational change in plectin that is inhibited by an engineered interdomain disulfide bridge. A two-step induced fit mechanism involving binding and subsequent domain rearrangement is proposed. In contrast, interaction with integrin alpha6beta4 occurs in a closed conformation. Competitive binding of plectin to F-actin and integrin alpha6beta4 may rely on the observed alternative binding mechanisms and involve both allosteric and steric factors.
Organizational Affiliation:
Program on Cell Adhesion, The Burnham Institute, 10901 North Torrey Pines Road, La Jolla, CA 92037, USA.