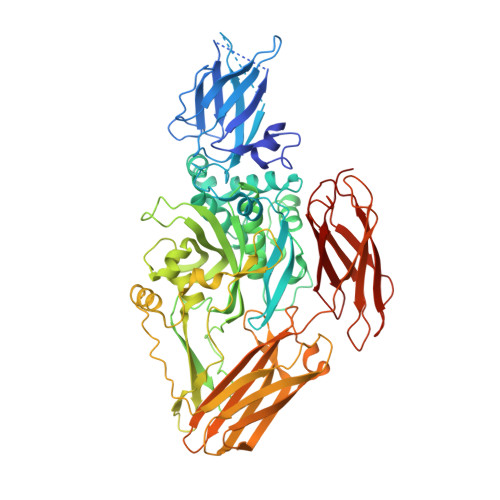
Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity.
Liu, S., Cerione, R.A., Clardy, J.(2002) Proc Natl Acad Sci U S A 99: 2743-2747
- PubMed: 11867708
- DOI: https://doi.org/10.1073/pnas.042454899
- Primary Citation of Related Structures:
1KV3 - PubMed Abstract:
Tissue transglutaminase (TG) is a Ca2+-dependent acyltransferase with roles in cellular differentiation, apoptosis, and other biological functions. In addition to being a transamidase, TG undergoes a GTP-binding/GTPase cycle even though it lacks any obvious sequence similarity with canonical GTP-binding (G) proteins. Guanine nucleotide binding and Ca2+ concentration reciprocally regulate TG's transamidation activity, with nucleotide binding being the negative regulator. Here we report the x-ray structure determined to 2.8-A resolution of human TG complexed with GDP. Although the transamidation active site is similar to those of other known transglutaminases, the guanine nucleotide-binding site of TG differs markedly from other G proteins. The structure suggests a structural basis for the negative regulation of transamidation activity by bound nucleotide, and the positive regulation of transamidation by Ca2+.
Organizational Affiliation:
Department of Chemistry, Cornell University, Ithaca, NY 14853, USA.