Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP K KH3 and single-stranded DNA.
Braddock, D.T., Baber, J.L., Levens, D., Clore, G.M.(2002) EMBO J 21: 3476-3485
- PubMed: 12093748
- DOI: https://doi.org/10.1093/emboj/cdf352
- Primary Citation of Related Structures:
1J5K - PubMed Abstract:
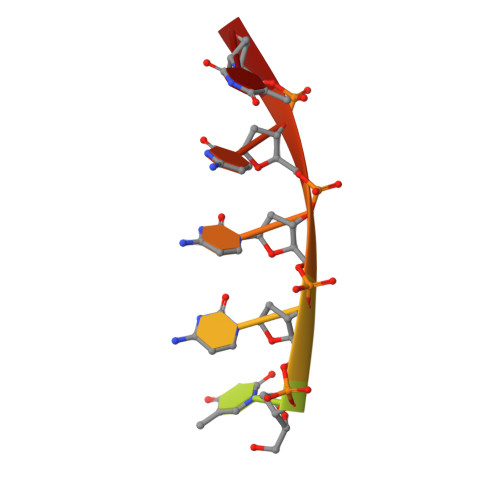
To elucidate the basis of sequence-specific single-stranded (ss) DNA recognition by K homology (KH) domains, we have solved the solution structure of a complex between the KH3 domain of the transcriptional regulator heterogeneous nuclear ribonucleoprotein K (hnRNP K) and a 10mer ssDNA. We show that hnRNP K KH3 specifically recognizes a tetrad of sequence 5'd-TCCC. The complex is stabilized by a dense network of methyl-oxygen hydrogen bonds involving the methyl groups of three isoleucine residues and the O2 and N3 atoms of the two central cytosine bases. Comparison with the recently solved structure of a specific protein-ssDNA complex involving the KH3 and KH4 domains of the far upstream element (FUSE) binding protein FBP suggests that the amino acid located five residues N-terminal of the invariant GXXG motif, which is characteristic of all KH domains, plays a crucial role in discrimination of the first two bases of the tetrad.
Organizational Affiliation:
Laboratory of Chemical Physics, Building 5, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892-0510, USA.