How the Pseudomonas Aeruginosa Exos Toxin Downregulates Rac
Wurtele, M., Wolf, E., Pederson, K.J., Buchwald, G., Ahmadian, M.R., Barbieri, J.T., Wittinghofer, A.(2001) Nat Struct Biol 8: 23
- PubMed: 11135665
- DOI: https://doi.org/10.1038/83007
- Primary Citation of Related Structures:
1HE1 - PubMed Abstract:
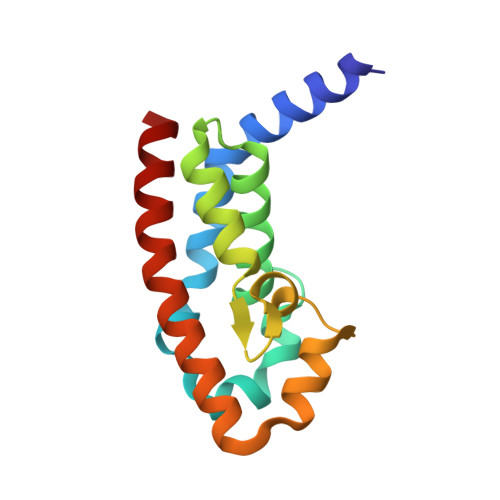
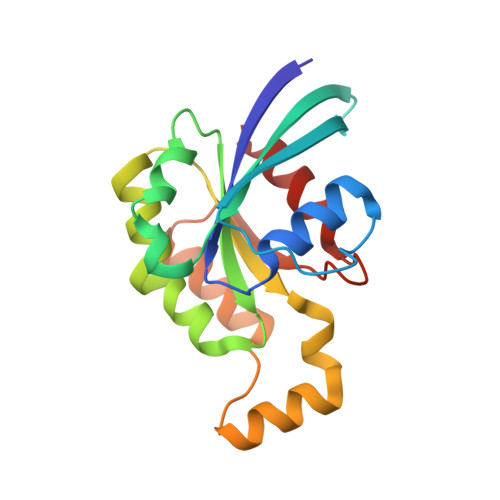
Pseudomonas aeruginosa is an opportunistic bacterial pathogen. One of its major toxins, ExoS, is translocated into eukaryotic cells by a type III secretion pathway. ExoS is a dual function enzyme that affects two different Ras-related GTP binding proteins. The C-terminus inactivates Ras through ADP ribosylation, while the N-terminus inactivates Rho proteins through its GTPase activating protein (GAP) activity. Here we have determined the three-dimensional structure of a complex between Rac and the GAP domain of ExoS in the presence of GDP and AlF3. Composed of approximately 130 residues, this ExoS domain is the smallest GAP hitherto described. The GAP domain of ExoS is an all-helical protein with no obvious structural homology, and thus no recognizable evolutionary relationship, with the eukaryotic RhoGAP or RasGAP fold. Similar to other GAPs, ExoS downregulates Rac using an arginine finger to stabilize the transition state of the GTPase reaction, but the details of the ExoS-Rac interaction are unique. Considering the intrinsic resistance of P. aeruginosa to antibiotics, this might open up a new avenue towards blocking its pathogenicity.
Organizational Affiliation:
Max-Planck-Institut für molekulare Physiologie, Abteilung Strukturelle Biologie, Otto-Hahn-Str. 11, 44227 Dortmund, Germany.