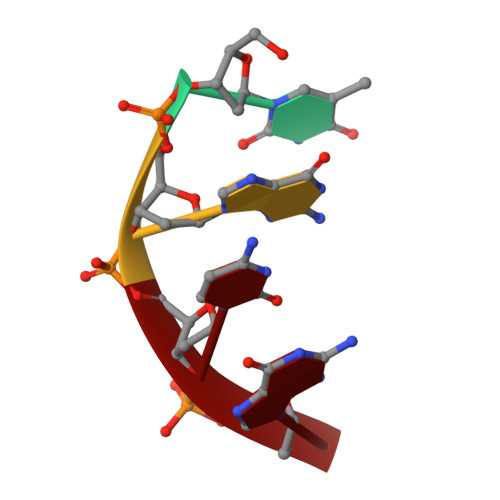
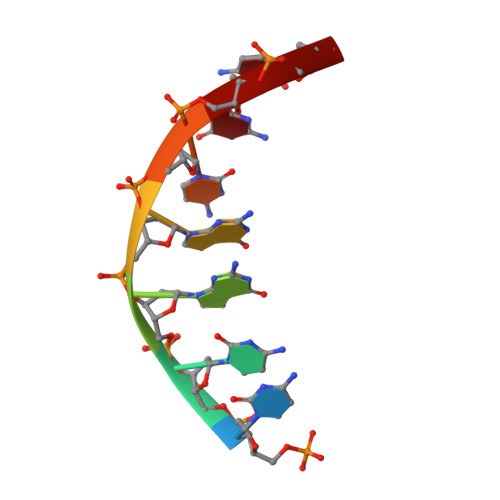
Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA.
Deibert, M., Grazulis, S., Sasnauskas, G., Siksnys, V., Huber, R.(2000) Nat Struct Biol 7: 792-799
- PubMed: 10966652
- DOI: https://doi.org/10.1038/79032
- Primary Citation of Related Structures:
1FIU - PubMed Abstract:
The crystal structure of the NgoMIV restriction endonuclease in complex with cleaved DNA has been determined at 1.6 A resolution. The crystallographic asymmetric unit contains a protein tetramer and two DNA molecules cleaved at their recognition sites. This is the first structure of a tetrameric restriction enzyme-DNA complex. In the tetramer, two primary dimers are arranged back to back with two oligonucleotides bound in clefts on opposite sides of the tetramer. The DNA molecules retain a B-type conformation and have an enclosed angle between their helical axes of 60 degrees. Sequence-specific interactions occur in both the major and minor grooves. Two Mg2+ ions are located close to the cleaved phosphate at the active site of NgoMIV. Biochemical experiments show that interactions between the recognition sites within the tetramer greatly increase DNA cleavage efficiency.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, D-82152 Planegg-Martinsried, Germany. markus.deibert@cii.de