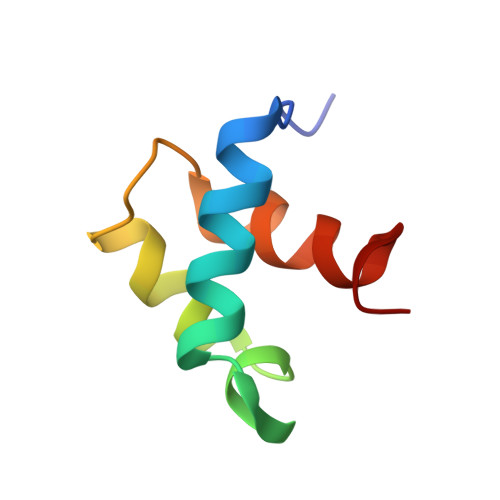
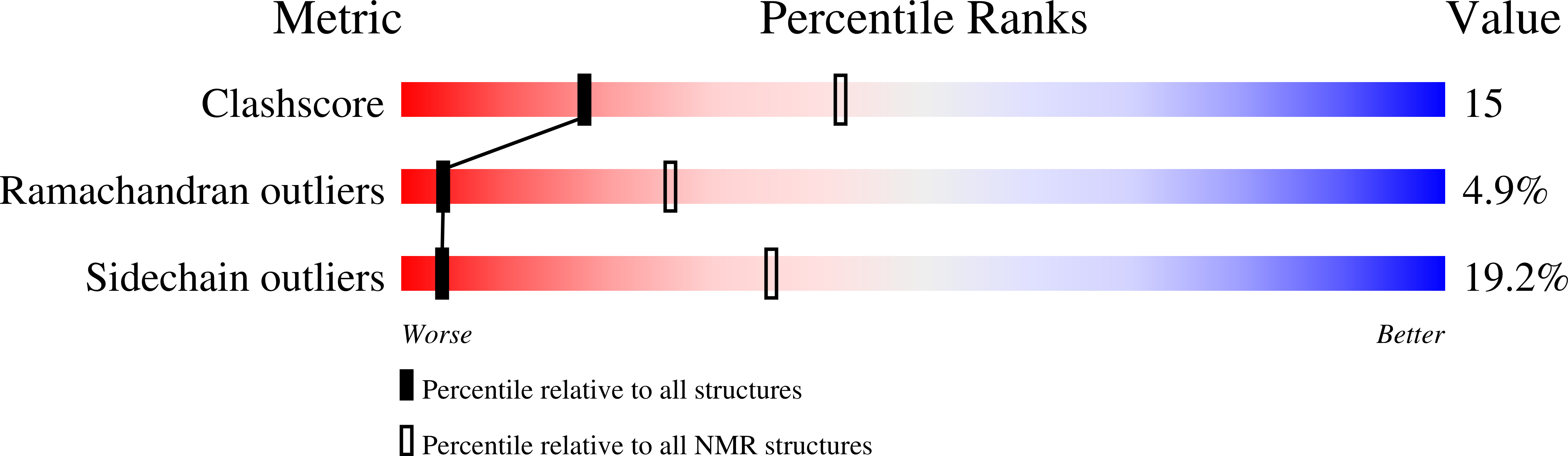NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains.
Hanaoka, S., Nagadoi, A., Yoshimura, S., Aimoto, S., Li, B., de Lange, T., Nishimura, Y.(2001) J Mol Biol 312: 167-175
- PubMed: 11545594
- DOI: https://doi.org/10.1006/jmbi.2001.4924
- Primary Citation of Related Structures:
1FEX - PubMed Abstract:
Mammalian telomeres are composed of long tandem arrays of double-stranded telomeric TTAGGG repeats associated with the telomeric DNA-binding proteins, TRF1 and TRF2. TRF1 and TRF2 contain a similar C-terminal Myb domain that mediates sequence-specific binding to telomeric DNA. In the budding yeast, telomeric DNA is associated with scRap1p, which has a central DNA-binding domain that contains two structurally related Myb domains connected by a long linker, an N-terminal BRCT domain, and a C-terminal RCT domain. Recently, the human ortholog of scRap1p (hRap1) was identified and shown to contain a BRCT domain and an RCT domain similar to scRap1p. However, hRap1 contained only one recognizable Myb motif in the center of the protein. Furthermore, while scRap1p binds telomeric DNA directly, hRap1 has no DNA-binding ability. Instead, hRap1 is tethered to telomeres by TRF2. Here, we have determined the solution structure of the Myb domain of hRap1 by NMR. It contains three helices maintained by a hydrophobic core. The architecture of the hRap1 Myb domain is very close to that of each of the Myb domains from TRF1, scRap1p and c-Myb. However, the electrostatic potential surface of the hRap1 Myb domain is distinguished from that of the other Myb domains. Each of the minimal DNA-binding domains, containing one Myb domain in TRF1 and two Myb domains in scRap1p and c-Myb, exhibits a positively charged broad surface that contacts closely the negatively charged backbone of DNA. By contrast, the hRap1 Myb domain shows no distinct positive surface, explaining its lack of DNA-binding activity. The hRap1 Myb domain may be a member of a second class of Myb motifs that lacks DNA-binding activity but may interact instead with other proteins. Other possible members of this class are the c-Myb R1 Myb domain and the Myb domains of ADA2 and Adf1. Thus, while the folds of all Myb domains resemble each other closely, the function of each Myb domain depends on the amino acid residues that are located on the surface of each protein.
Organizational Affiliation:
Graduate School of Integrated Science, Yokohama City University, 1-7-29 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan.