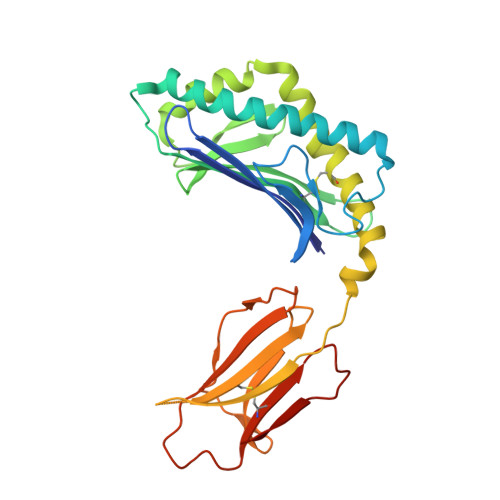
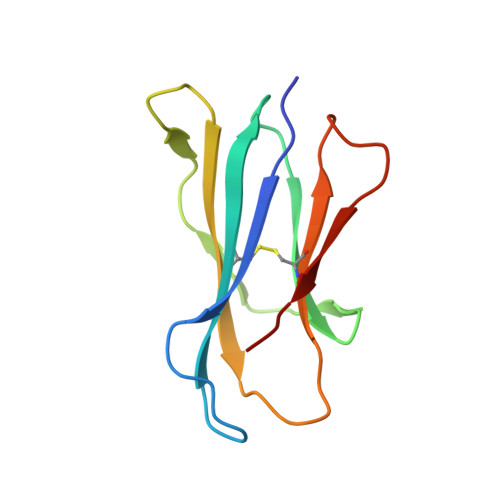
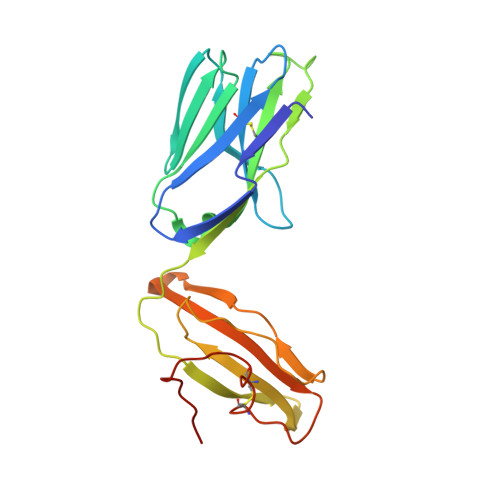
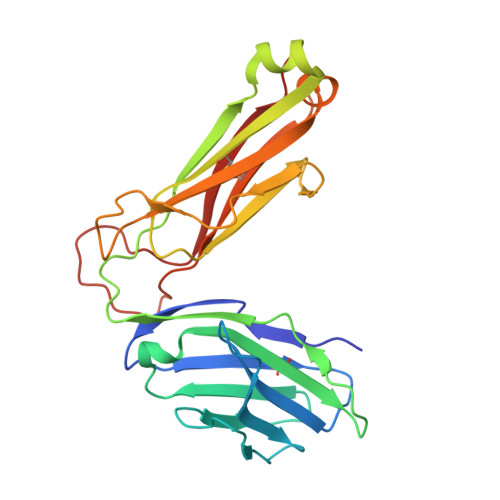
Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Aspeslagh, S., Li, Y., Yu, E.D., Pauwels, N., Trappeniers, M., Girardi, E., Decruy, T., Van Beneden, K., Venken, K., Drennan, M., Leybaert, L., Wang, J., Franck, R.W., Van Calenbergh, S., Zajonc, D.M., Elewaut, D.(2011) EMBO J 30: 2294-2305
- PubMed: 21552205
- DOI: https://doi.org/10.1038/emboj.2011.145
- Primary Citation of Related Structures:
3QUX, 3QUY, 3QUZ - PubMed Abstract:
Invariant natural killer T (iNKT) cells are known to have marked immunomodulatory capacity due to their ability to produce copious amounts of effector cytokines. Here, we report the structure and function of a novel class of aromatic α-galactosylceramide structurally related glycolipids with marked Th1 bias in both mice and men, leading to superior tumour protection in vivo. The strength of the Th1 response correlates well with enhanced lipid binding to CD1d as a result of an induced fit mechanism that binds the aromatic substitution as a third anchor, in addition to the two lipid chains. This induced fit is in contrast to another Th1-biasing glycolipid, α-C-GalCer, whose CD1d binding follows a conventional key-lock principle. These findings highlight the previously unexploited flexibility of CD1d in accommodating galactose-modified glycolipids and broaden the range of glycolipids that can stimulate iNKT cells. We speculate that glycolipids can be designed that induce a similar fit, thereby leading to superior and more sustained iNKT cell responses in vivo.
Organizational Affiliation:
Department of Rheumatology, Faculty of Medicine and Health Sciences, Laboratory for Molecular Immunology and Inflammation, Ghent University, Ghent, Belgium.