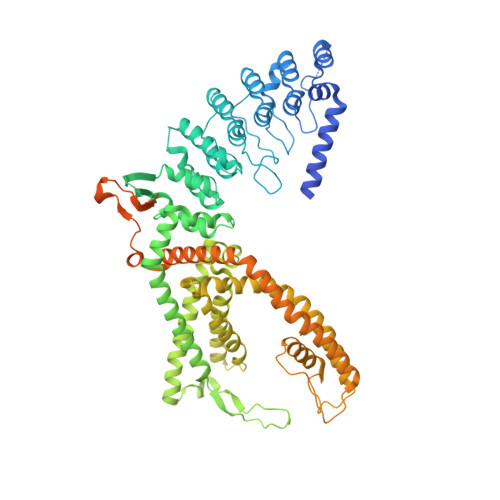
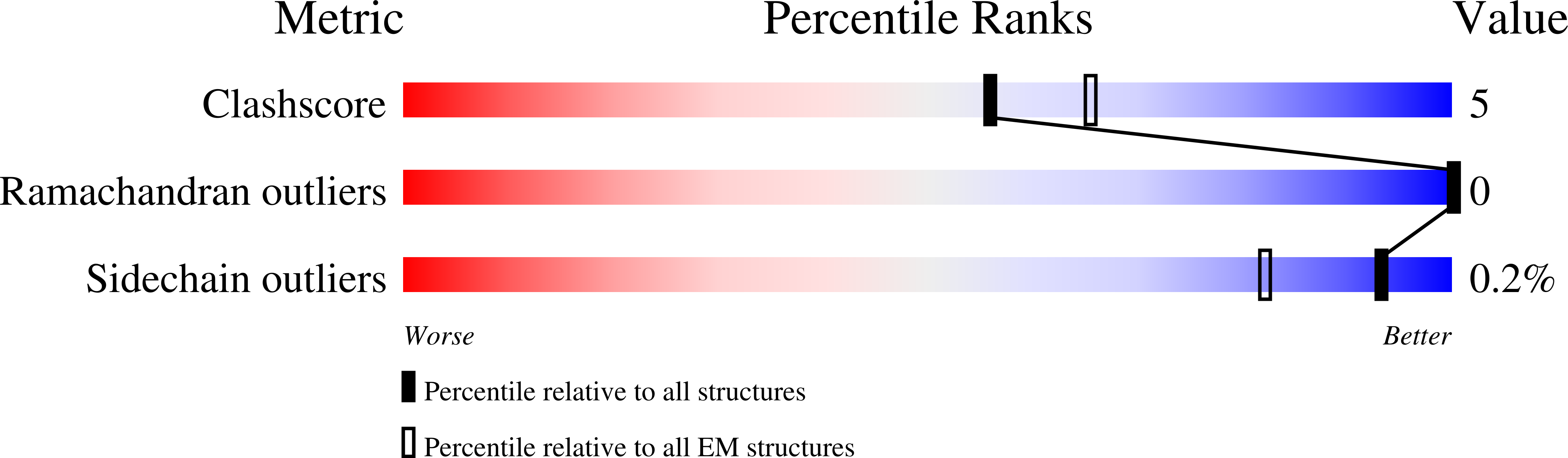Structural mechanism of TRPV5 inhibition by econazole.
De Jesus-Perez, J.J., Gabrielle, M., Raheem, S., Fluck, E.C., Rohacs, T., Moiseenkova-Bell, V.Y.(2024) Structure 32: 148-156.e5
- PubMed: 38141613
- DOI: https://doi.org/10.1016/j.str.2023.11.012
- Primary Citation of Related Structures:
8TF2, 8TF3, 8TF4 - PubMed Abstract:
The calcium-selective TRPV5 channel activated by phosphatidylinositol 4,5-bisphosphate [PI(4,5)P 2 ] is involved in calcium homeostasis. Recently, cryoelectron microscopy (cryo-EM) provided molecular details of TRPV5 modulation by exogenous and endogenous molecules. However, the details of TRPV5 inhibition by the antifungal agent econazole (ECN) remain elusive due to the low resolution of the currently available structure. In this study, we employ cryo-EM to comprehensively examine how the ECN inhibits TRPV5. By combining our structural findings with site-directed mutagenesis, calcium measurements, electrophysiology, and molecular dynamics simulations, we determined that residues F472 and L475 on the S4 helix, along with residue W495 on the S5 helix, collectively constitute the ECN-binding site. Additionally, the structure of TRPV5 in the presence of ECN and PI(4,5)P 2 , which does not show the bound activator, reveals a potential inhibition mechanism in which ECN competes with PI(4,5)P 2 , preventing the latter from binding, and ultimately pore closure.
Organizational Affiliation:
Department of Systems Pharmacology and Translational Therapeutics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.