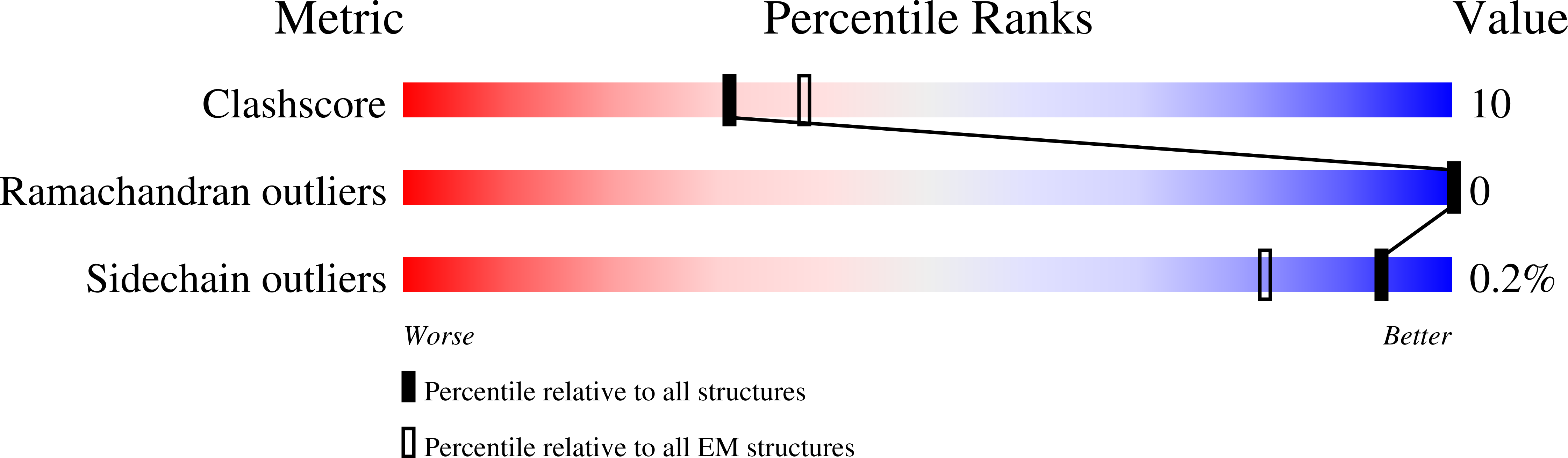Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Moseng, M.A., Su, C.C., Rios, K., Cui, M., Lyu, M., Glaza, P., Klenotic, P.A., Delpire, E., Yu, E.W.(2022) Sci Adv 8: eabq0952
- PubMed: 36306358
- DOI: https://doi.org/10.1126/sciadv.abq0952
- Primary Citation of Related Structures:
7MXO, 7N3N, 7SFL, 7SMP, 8STE - PubMed Abstract:
The Na-K-2Cl cotransporter-1 (NKCC1) is an electroneutral Na + -dependent transporter responsible for simultaneously translocating Na + , K + , and Cl - ions into cells. In human tissue, NKCC1 plays a critical role in regulating cytoplasmic volume, fluid intake, chloride homeostasis, and cell polarity. Here, we report four structures of human NKCC1 (hNKCC1), both in the absence and presence of loop diuretic (bumetanide or furosemide), using single-particle cryo-electron microscopy. These structures allow us to directly observe various novel conformations of the hNKCC1 dimer. They also reveal two drug-binding sites located at the transmembrane and cytosolic carboxyl-terminal domains, respectively. Together, our findings enable us to delineate an inhibition mechanism that involves a coupled movement between the cytosolic and transmembrane domains of hNKCC1.
Organizational Affiliation:
Department of Pharmacology, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA.