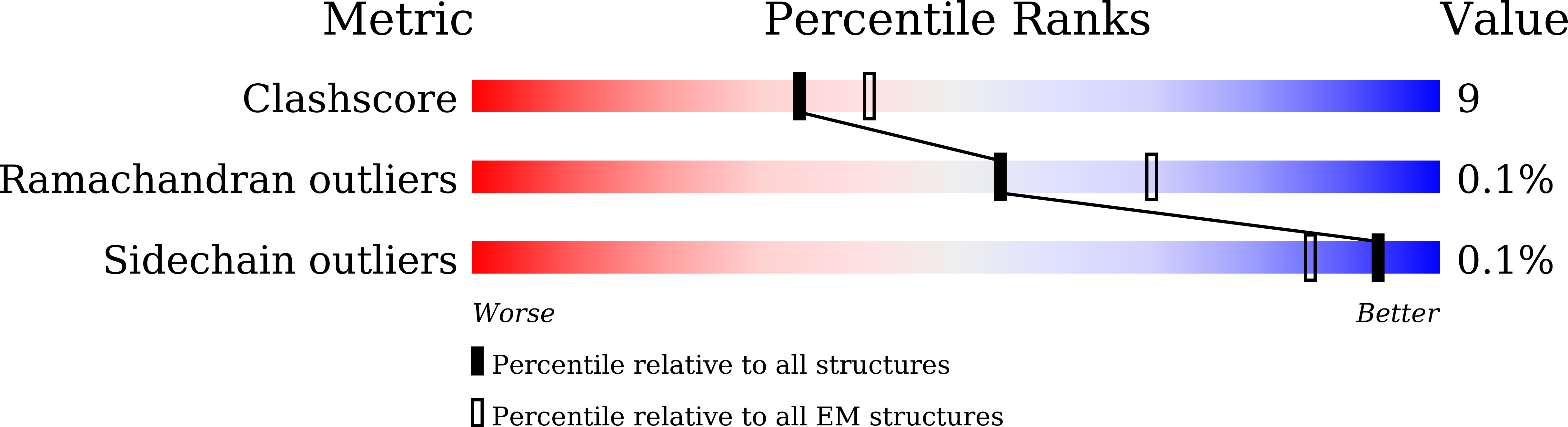Recognition Mechanism of a Novel Gabapentinoid Drug, Mirogabalin, for Recombinant Human alpha 2 delta 1, a Voltage-Gated Calcium Channel Subunit.
Kozai, D., Numoto, N., Nishikawa, K., Kamegawa, A., Kawasaki, S., Hiroaki, Y., Irie, K., Oshima, A., Hanzawa, H., Shimada, K., Kitano, Y., Fujiyoshi, Y.(2023) J Mol Biol 435: 168049-168049
- PubMed: 36933823
- DOI: https://doi.org/10.1016/j.jmb.2023.168049
- Primary Citation of Related Structures:
8IF3, 8IF4 - PubMed Abstract:
Mirogabalin is a novel gabapentinoid drug with a hydrophobic bicyclo substituent on the γ-aminobutyric acid moiety that targets the voltage-gated calcium channel subunit α 2 δ1. Here, to reveal the mirogabalin recognition mechanisms of α 2 δ1, we present structures of recombinant human α 2 δ1 with and without mirogabalin analyzed by cryo-electron microscopy. These structures show the binding of mirogabalin to the previously reported gabapentinoid binding site, which is the extracellular dCache_1 domain containing a conserved amino acid binding motif. A slight conformational change occurs around the residues positioned close to the hydrophobic group of mirogabalin. Mutagenesis binding assays identified that residues in the hydrophobic interaction region, in addition to several amino acid binding motif residues around the amino and carboxyl groups of mirogabalin, are critical for mirogabalin binding. The A215L mutation introduced to decrease the hydrophobic pocket volume predictably suppressed mirogabalin binding and promoted the binding of another ligand, L-Leu, with a smaller hydrophobic substituent than mirogabalin. Alterations of residues in the hydrophobic interaction region of α 2 δ1 to those of the α 2 δ2, α 2 δ3, and α 2 δ4 isoforms, of which α 2 δ3 and α 2 δ4 are gabapentin-insensitive, suppressed the binding of mirogabalin. These results support the importance of hydrophobic interactions in α 2 δ1 ligand recognition.
Organizational Affiliation:
Cellular and Structural Physiology Institute (CeSPI), Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8601, Japan; Japan Biological Informatics Consortium, 2-4-32 Aomi, Koto-ku, Tokyo 135-0063, Japan; Advanced Research Institute, Tokyo Medical and Dental University, 1-5-45 Yushima, Bunkyo-ku, Tokyo 113-8501, Japan.