An ependymin-related blue carotenoprotein decorates marine blue sponge.
Kawasaki, S., Kaneko, T., Asano, T., Maoka, T., Takaichi, S., Shomura, Y.(2023) J Biol Chem 299: 105110-105110
- PubMed: 37517696
- DOI: https://doi.org/10.1016/j.jbc.2023.105110
- Primary Citation of Related Structures:
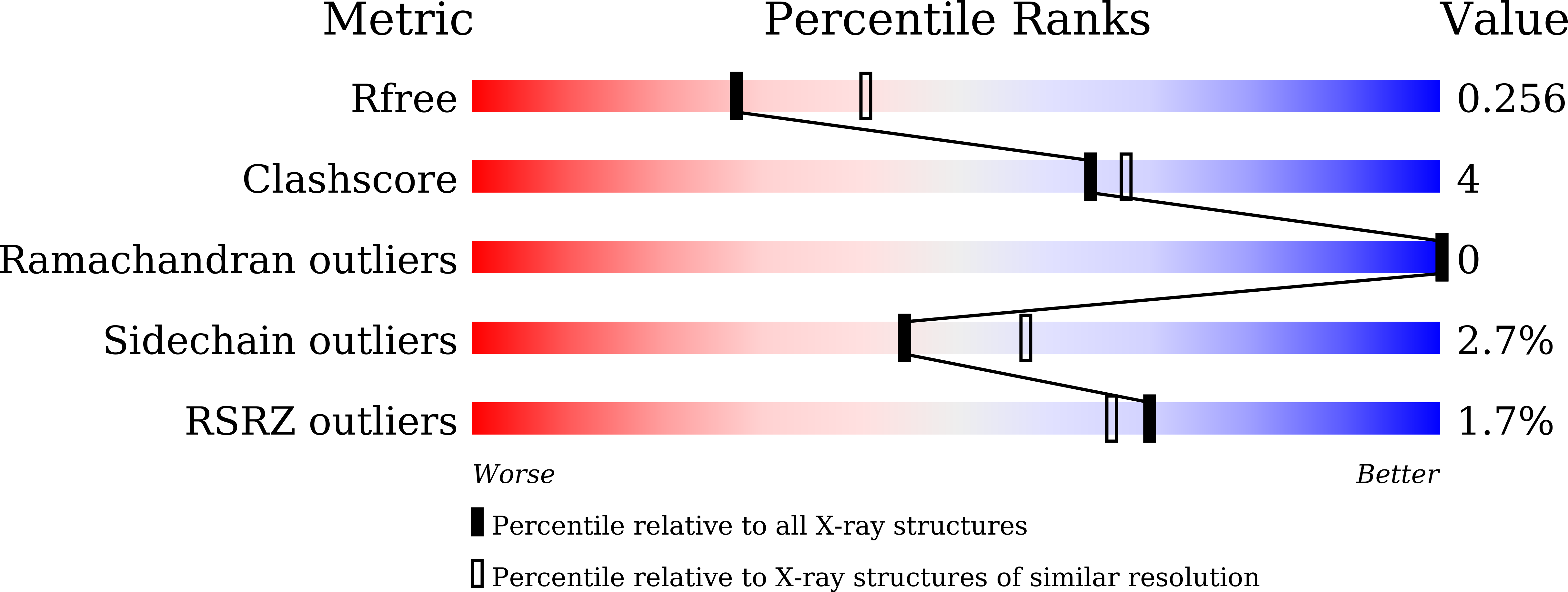
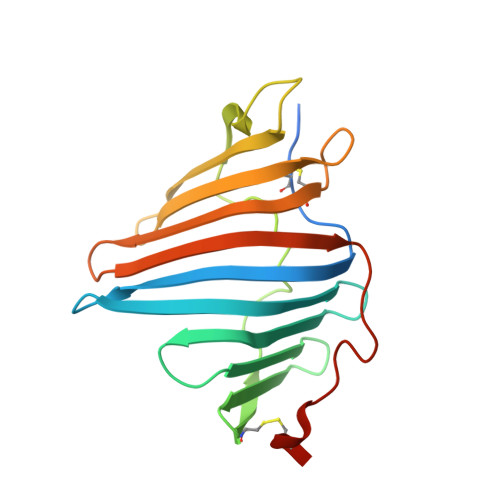
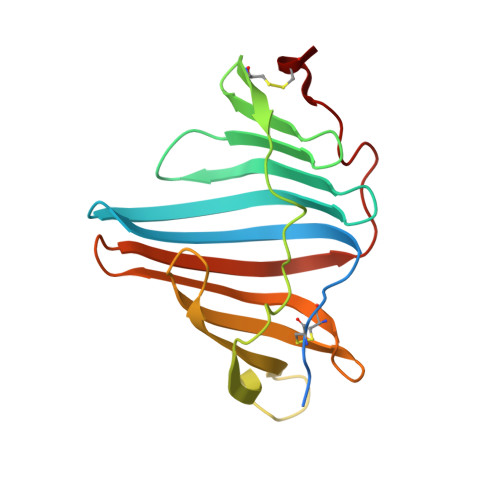
8I34 - PubMed Abstract:
Marine animals display diverse vibrant colors, but the mechanisms underlying their specific coloration remain to be clarified. Blue coloration is known to be achieved through a bathochromic shift of the orange carotenoid astaxanthin (AXT) by the crustacean protein crustacyanin, but other examples have not yet been well investigated. Here, we identified an ependymin (EPD)-related water-soluble blue carotenoprotein responsible for the specific coloration of the marine blue sponge Haliclona sp. EPD was originally identified in the fish brain as a protein involved in memory consolidation and neuronal regeneration. The purified blue protein, designated as EPD-related blue carotenoprotein-1, was identified as a secreted glycoprotein. We show that it consists of a heterodimer that binds orange AXT and mytiloxanthin and exhibits a bathochromic shift. Our crystal structure analysis of the natively purified EPD-related blue carotenoprotein-1 revealed that these two carotenoids are specifically bound to the heterodimer interface, where the polyene chains are aligned in parallel to each other like in β-crustacyanin, although the two proteins are evolutionary and structurally unrelated. Furthermore, using reconstitution assays, we found that incomplete bathochromic shifts occurred when the protein bound to only AXT or mytiloxanthin. Taken together, we identified an EPD in a basal metazoan as a blue protein that decorates the sponge body by binding specific structurally unrelated carotenoids.
Organizational Affiliation:
Department of Molecular Microbiology, Tokyo University of Agriculture, Tokyo, Japan. Electronic address: kawashin@nodai.ac.jp.