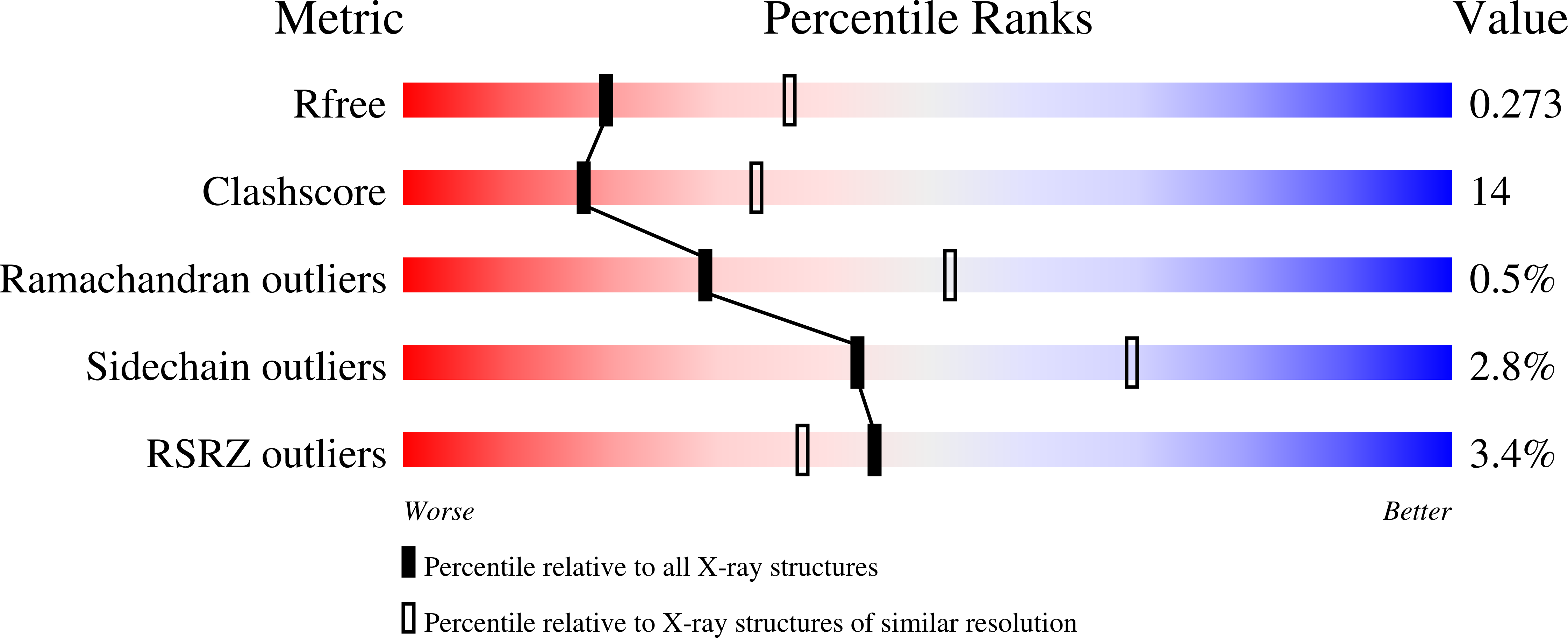
Human cytochrome P450 3A7 binding four copies of its native substrate dehydroepiandrosterone 3-sulfate.
Liu, J., Kandel, S.E., Lampe, J.N., Scott, E.E.(2023) J Biol Chem 299: 104993-104993
- PubMed: 37392852
- DOI: https://doi.org/10.1016/j.jbc.2023.104993
- Primary Citation of Related Structures:
8GK3 - PubMed Abstract:
Human fetal cytochrome P450 3A7 (CYP3A7) is involved in both xenobiotic metabolism and the estriol biosynthetic pathway. Although much is understood about cytochrome P450 3A4 and its role in adult drug metabolism, CYP3A7 is poorly characterized in terms of its interactions with both categories of substrates. Herein, a crystallizable mutated form of CYP3A7 was saturated with its primary endogenous substrate dehydroepiandrosterone 3-sulfate (DHEA-S) to yield a 2.6 Å X-ray structure revealing the unexpected capacity to simultaneously bind four copies of DHEA-S. Two DHEA-S molecules are located in the active site proper, one in a ligand access channel, and one on the hydrophobic F'-G' surface normally embedded in the membrane. While neither DHEA-S binding nor metabolism exhibit cooperative kinetics, the current structure is consistent with cooperativity common to CYP3A enzymes. Overall, this information suggests that mechanism(s) of CYP3A7 interactions with steroidal substrates are complex.
Organizational Affiliation:
Department of Medicinal Chemistry, University of Michigan, Ann Arbor, Michigan, USA.