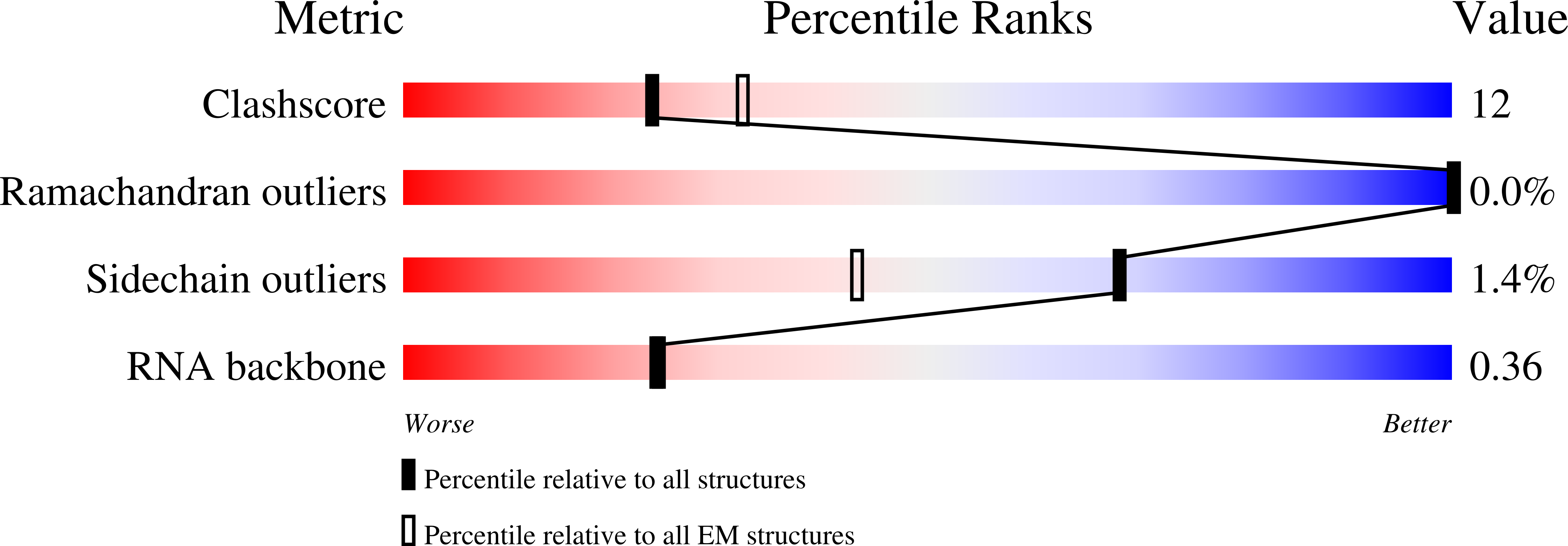
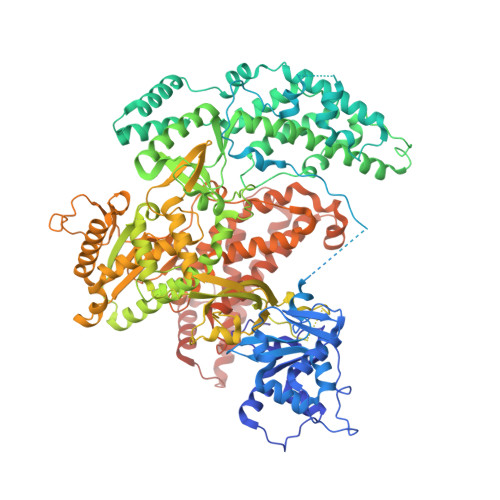
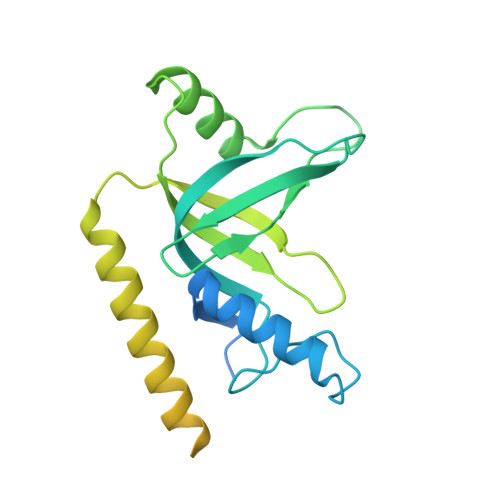
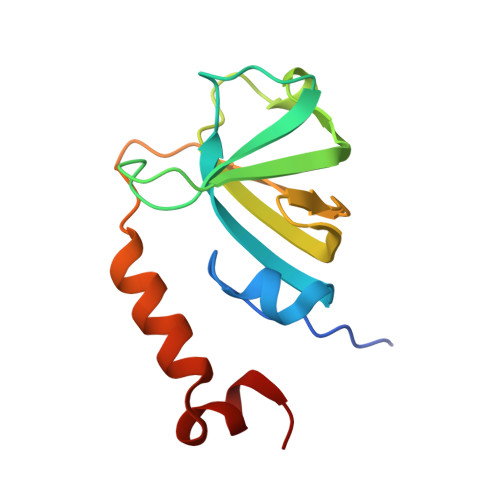
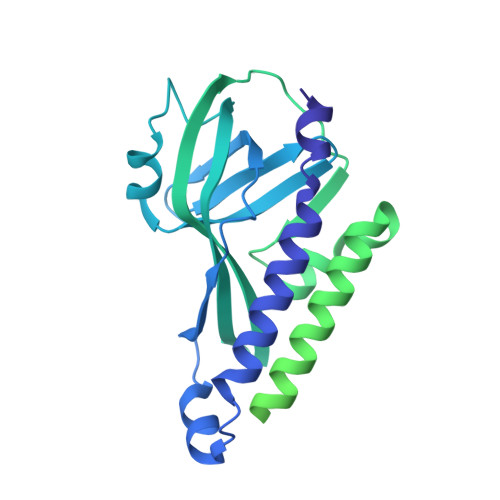
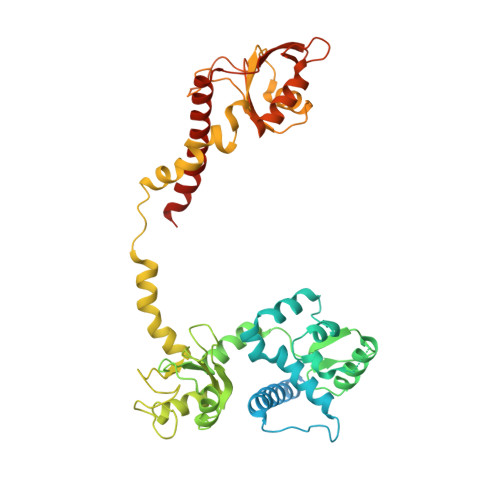
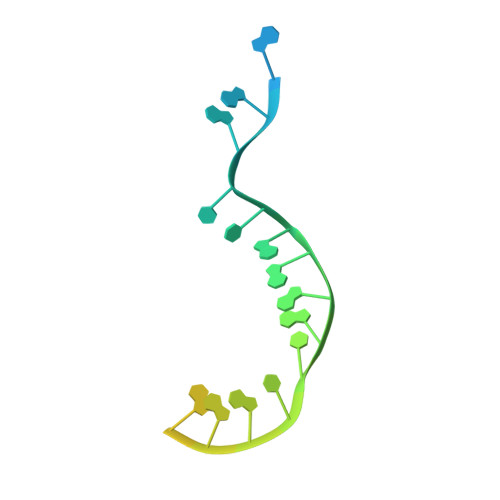
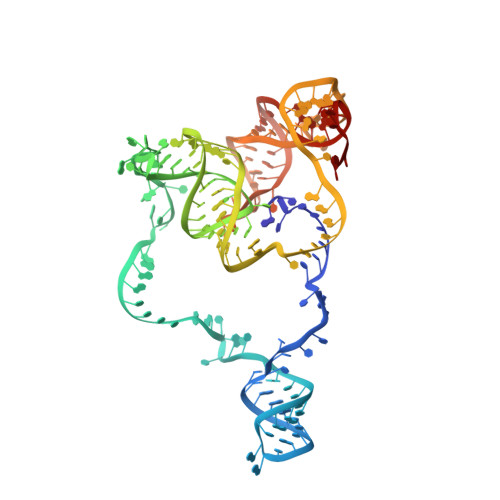
Structure of LARP7 Protein p65-telomerase RNA Complex in Telomerase Revealed by Cryo-EM and NMR.
Wang, Y., He, Y., Wang, Y., Yang, Y., Singh, M., Eichhorn, C.D., Cheng, X., Jiang, Y.X., Zhou, Z.H., Feigon, J.(2023) J Mol Biol 435: 168044-168044
- PubMed: 37330293
- DOI: https://doi.org/10.1016/j.jmb.2023.168044
- Primary Citation of Related Structures:
8GAP - PubMed Abstract:
La-related protein 7 (LARP7) are a family of RNA chaperones that protect the 3'-end of RNA and are components of specific ribonucleoprotein complexes (RNP). In Tetrahymena thermophila telomerase, LARP7 protein p65 together with telomerase reverse transcriptase (TERT) and telomerase RNA (TER) form the core RNP. p65 has four known domains-N-terminal domain (NTD), La motif (LaM), RNA recognition motif 1 (RRM1), and C-terminal xRRM2. To date, only the xRRM2 and LaM and their interactions with TER have been structurally characterized. Conformational dynamics leading to low resolution in cryo-EM density maps have limited our understanding of how full-length p65 specifically recognizes and remodels TER for telomerase assembly. Here, we combined focused classification of Tetrahymena telomerase cryo-EM maps with NMR spectroscopy to determine the structure of p65-TER. Three previously unknown helices are identified, one in the otherwise intrinsically disordered NTD that binds the La module, one that extends RRM1, and another preceding xRRM2, that stabilize p65-TER interactions. The extended La module (αN, LaM and RRM1) interacts with the four 3' terminal U nucleotides, while LaM and αN additionally interact with TER pseudoknot, and LaM with stem 1 and 5' end. Our results reveal the extensive p65-TER interactions that promote TER 3'-end protection, TER folding, and core RNP assembly and stabilization. The structure of full-length p65 with TER also sheds light on the biological roles of genuine La and LARP7 proteins as RNA chaperones and core RNP components.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California Los Angeles, Los Angeles, CA 90095-1569, USA.