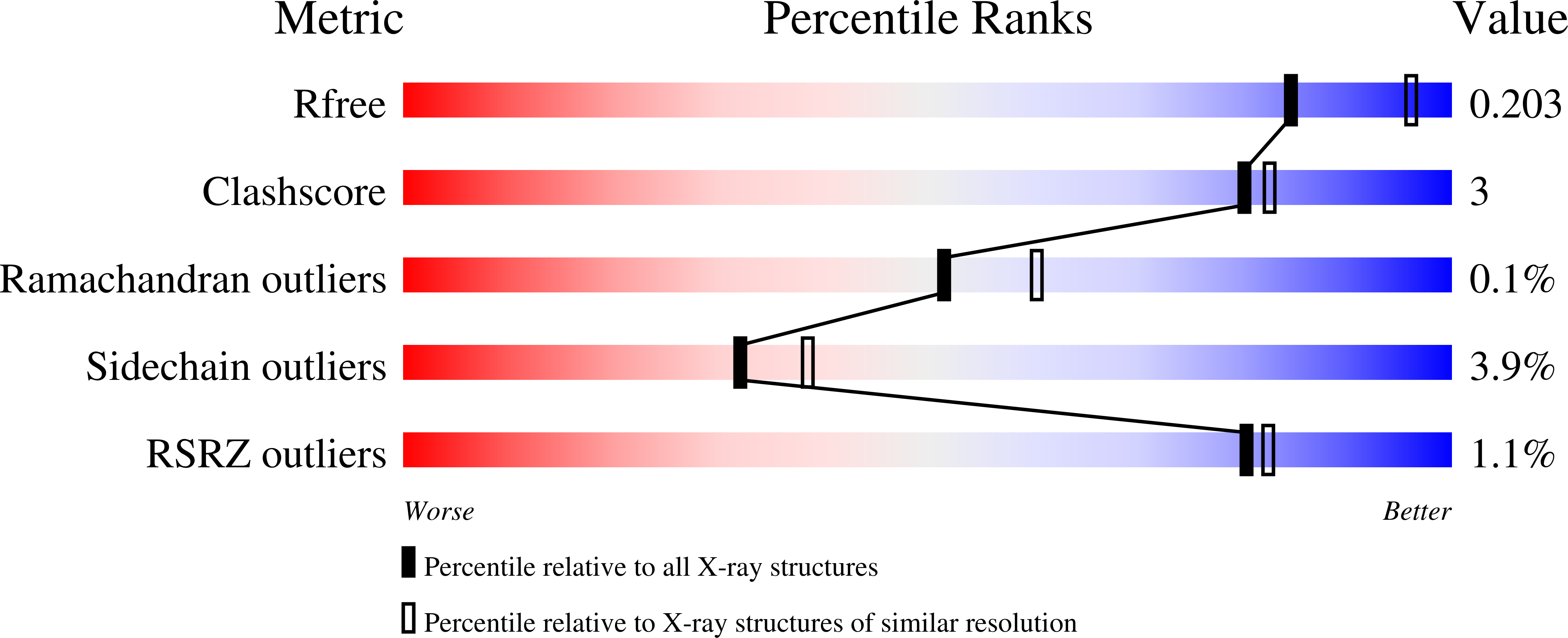
Multidisciplinary docking, kinetics and X-ray crystallography studies of baicalein acting as a glycogen phosphorylase inhibitor and determination of its' potential against glioblastoma in cellular models.
Mathomes, R.T., Koulas, S.M., Tsialtas, I., Stravodimos, G., Welsby, P.J., Psarra, A.G., Stasik, I., Leonidas, D.D., Hayes, J.M.(2023) Chem Biol Interact 382: 110568-110568
- PubMed: 37277066
- DOI: https://doi.org/10.1016/j.cbi.2023.110568
- Primary Citation of Related Structures:
8BZS - PubMed Abstract:
Glycogen phosphorylase (GP) is the rate-determining enzyme in the glycogenolysis pathway. Glioblastoma (GBM) is amongst the most aggressive cancers of the central nervous system. The role of GP and glycogen metabolism in the context of cancer cell metabolic reprogramming is recognised, so that GP inhibitors may have potential treatment benefits. Here, baicalein (5,6,7-trihydroxyflavone) is studied as a GP inhibitor, and for its effects on glycogenolysis and GBM at the cellular level. The compound is revealed as a potent GP inhibitor against human brain GPa (K i = 32.54 μM), human liver GPa (K i = 8.77 μM) and rabbit muscle GPb (K i = 5.66 μM) isoforms. It is also an effective inhibitor of glycogenolysis (IC 50 = 119.6 μM), measured in HepG2 cells. Most significantly, baicalein demonstrated anti-cancer potential through concentration- and time-dependent decrease in cell viability for three GBM cell-lines (U-251 MG, U-87 MG, T98-G) with IC 50 values of ∼20-55 μM (48- and 72-h). Its effectiveness against T98-G suggests potential against GBM with resistance to temozolomide (the first-line therapy) due to a positive O 6 -methylguanine-DNA methyltransferase (MGMT) status. The solved X-ray structure of rabbit muscle GP-baicalein complex will facilitate structure-based design of GP inhibitors. Further exploration of baicalein and other GP inhibitors with different isoform specificities against GBM is suggested.
Organizational Affiliation:
School of Pharmacy & Biomedical Sciences, University of Central Lancashire, Preston, PR1 2HE, United Kingdom.