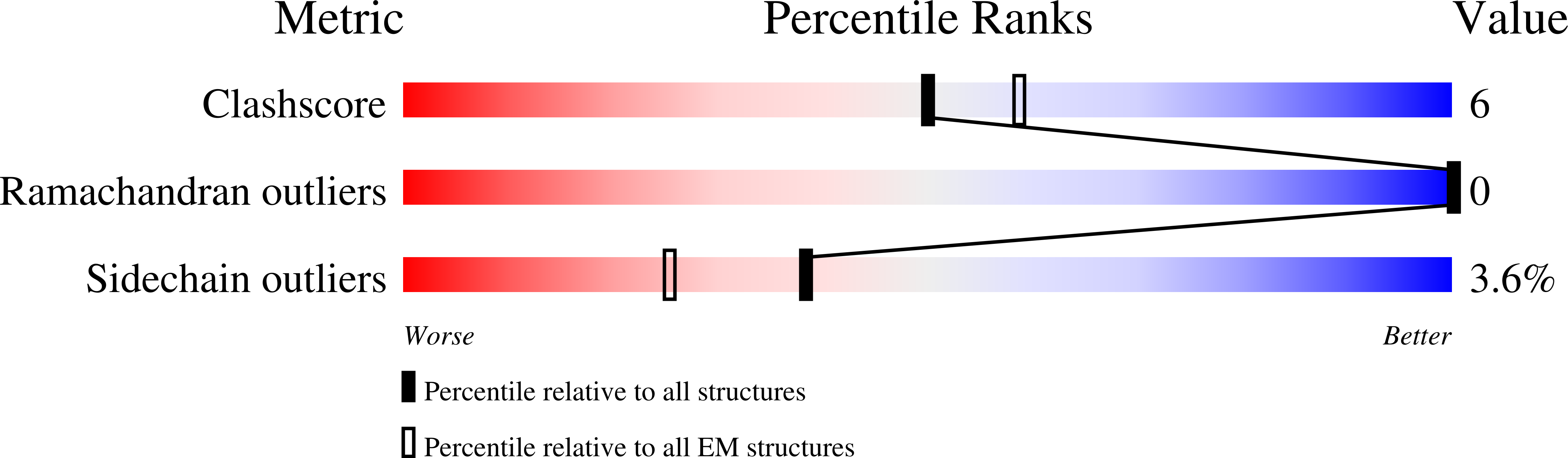Mechanism of RNA polymerase I selection by transcription factor UAF.
Baudin, F., Murciano, B., Fung, H.K.H., Fromm, S.A., Mattei, S., Mahamid, J., Muller, C.W.(2022) Sci Adv 8: eabn5725-eabn5725
- PubMed: 35442737
- DOI: https://doi.org/10.1126/sciadv.abn5725
- Primary Citation of Related Structures:
7Z0O - PubMed Abstract:
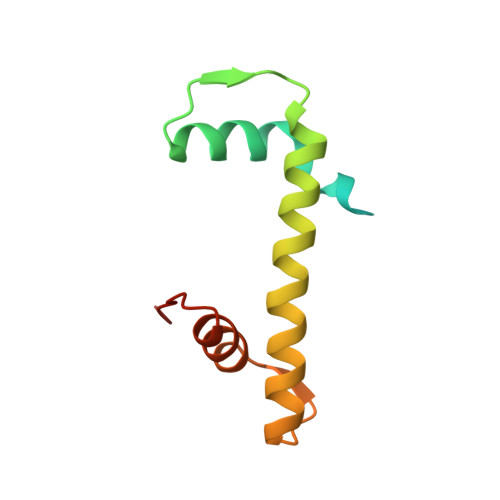
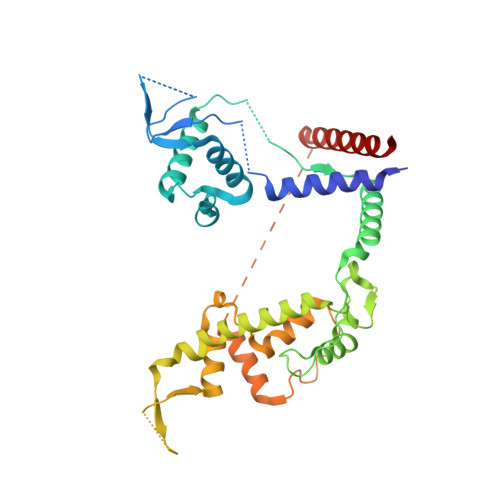
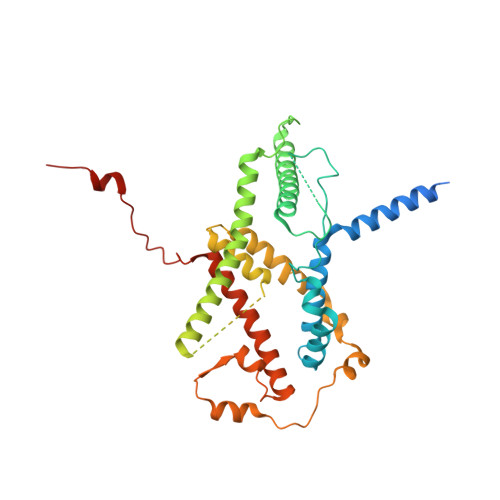
Preribosomal RNA is selectively transcribed by RNA polymerase (Pol) I in eukaryotes. The yeast transcription factor upstream activating factor (UAF) represses Pol II transcription and mediates Pol I preinitiation complex (PIC) formation at the 35 S ribosomal RNA gene. To visualize the molecular intermediates toward PIC formation, we determined the structure of UAF in complex with native promoter DNA and transcription factor TATA-box-binding protein (TBP). We found that UAF recognizes DNA using a hexameric histone-like scaffold with markedly different interactions compared with the nucleosome and the histone-fold-rich transcription factor IID (TFIID). In parallel, UAF positions TBP for Core Factor binding, which leads to Pol I recruitment, while sequestering it from DNA and Pol II/III-specific transcription factors. Our work thus reveals the structural basis of RNA Pol selection by a transcription factor.
Organizational Affiliation:
Structural and Computational Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany.