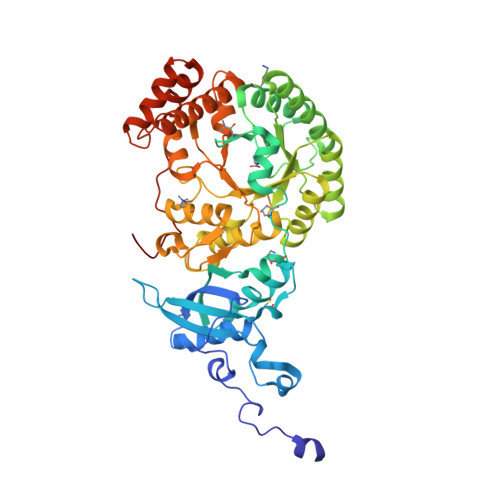
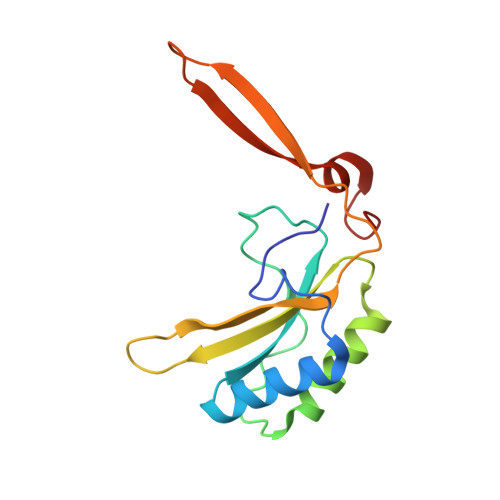
A linker protein from a red-type pyrenoid phase separates with Rubisco via oligomerizing sticker motifs.
Oh, Z.G., Ang, W.S.L., Poh, C.W., Lai, S.K., Sze, S.K., Li, H.Y., Bhushan, S., Wunder, T., Mueller-Cajar, O.(2023) Proc Natl Acad Sci U S A 120: e2304833120-e2304833120
- PubMed: 37311001
- DOI: https://doi.org/10.1073/pnas.2304833120
- Primary Citation of Related Structures:
7YK5 - PubMed Abstract:
The slow kinetics and poor substrate specificity of the key photosynthetic CO 2 -fixing enzyme Rubisco have prompted the repeated evolution of Rubisco-containing biomolecular condensates known as pyrenoids in the majority of eukaryotic microalgae. Diatoms dominate marine photosynthesis, but the interactions underlying their pyrenoids are unknown. Here, we identify and characterize the Rubisco linker protein PYCO1 from Phaeodactylum tricornutum . PYCO1 is a tandem repeat protein containing prion-like domains that localizes to the pyrenoid. It undergoes homotypic liquid-liquid phase separation (LLPS) to form condensates that specifically partition diatom Rubisco. Saturation of PYCO1 condensates with Rubisco greatly reduces the mobility of droplet components. Cryo-electron microscopy and mutagenesis data revealed the sticker motifs required for homotypic and heterotypic phase separation. Our data indicate that the PYCO1-Rubisco network is cross-linked by PYCO1 stickers that oligomerize to bind to the small subunits lining the central solvent channel of the Rubisco holoenzyme. A second sticker motif binds to the large subunit. Pyrenoidal Rubisco condensates are highly diverse and tractable models of functional LLPS.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, Singapore 637551, Singapore.