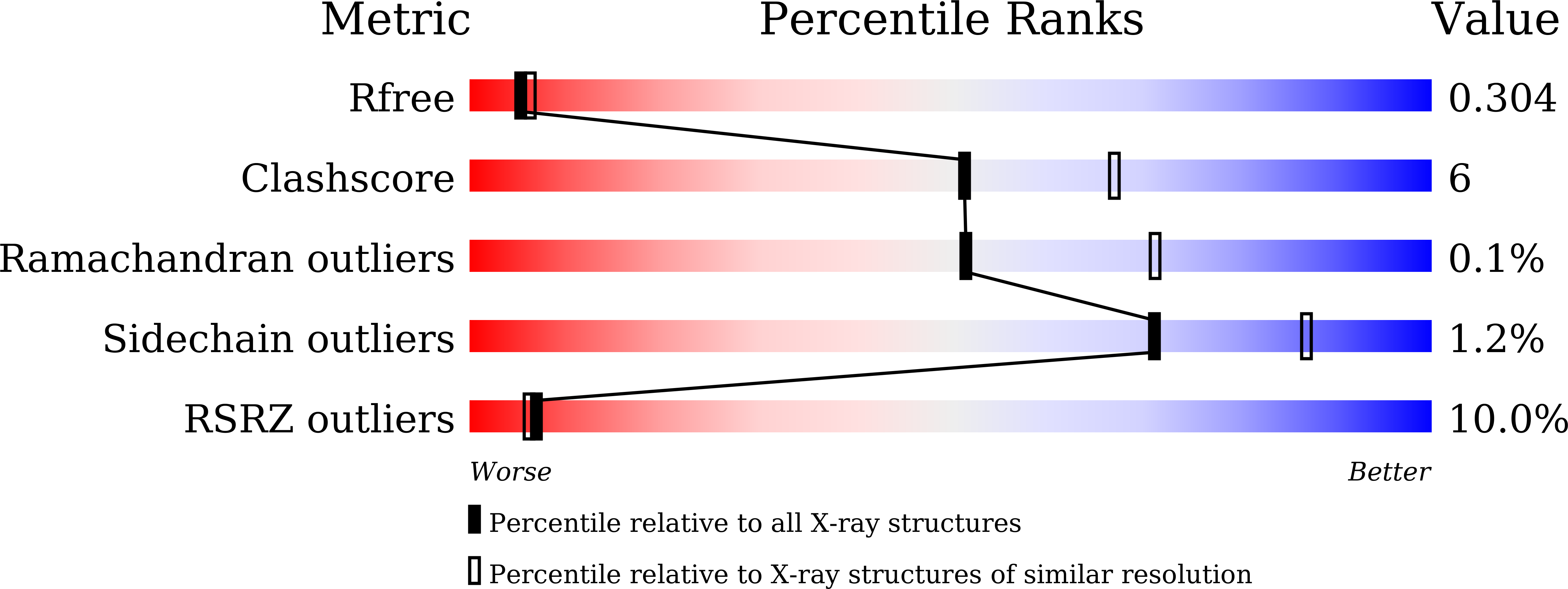
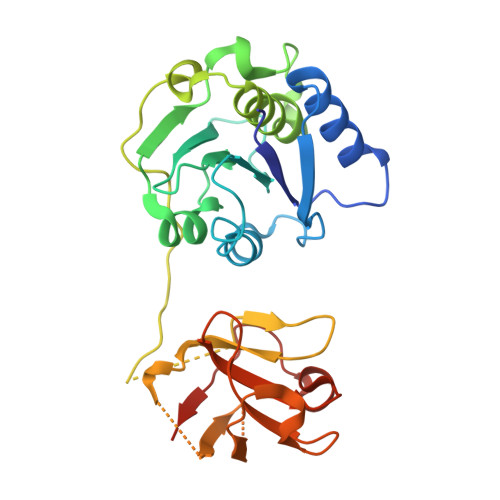
Expression, purification and structure determination of the chlorinase ClA2.
Liu, Y., Zhang, H., Xiao, H., Li, Y., Liu, Y.(2022) Biochem Biophys Res Commun 628: 64-67
- PubMed: 36081280
- DOI: https://doi.org/10.1016/j.bbrc.2022.08.081
- Primary Citation of Related Structures:
7XTO - PubMed Abstract:
Halogenated compounds are particularly important in pharmaceutical and agrochemical products. Biochemical halogenation with halogenases are environmental friendly reactions with high efficiency. Recently, the two new chlorinases (ClA1 and ClA2) were discovered from soil bacteria. However, the protein structure of ClA2 was not identified. Here, we determined the high-resolution crystal structure of ClA2. This structure will help us to understand the catalytic mechanism of chlorinase, and explain the catalytic process of the coupled chlorinase-fluorinase system, which offers the prospect of arising rapid radiolabeling protocols under mild conditions.
Organizational Affiliation:
Department of Pathogen Biology, School of Basic Medicine, Tongji Medical College, Huazhong University of Science and Technology, 13 Hangkong Road, Wuhan, Hubei, 430030, China.