Discovery of a Unique Structural Motif in Lanthipeptide Synthetases for Substrate Binding and Interdomain Interactions.
Huang, S., Wang, Y., Cai, C., Xiao, X., Liu, S., Ma, Y., Xie, X., Liang, Y., Chen, H., Zhu, J., Hegemann, J.D., Yao, H., Wei, W., Wang, H.(2022) Angew Chem Int Ed Engl 61: e202211382-e202211382
- PubMed: 36102578
- DOI: https://doi.org/10.1002/anie.202211382
- Primary Citation of Related Structures:
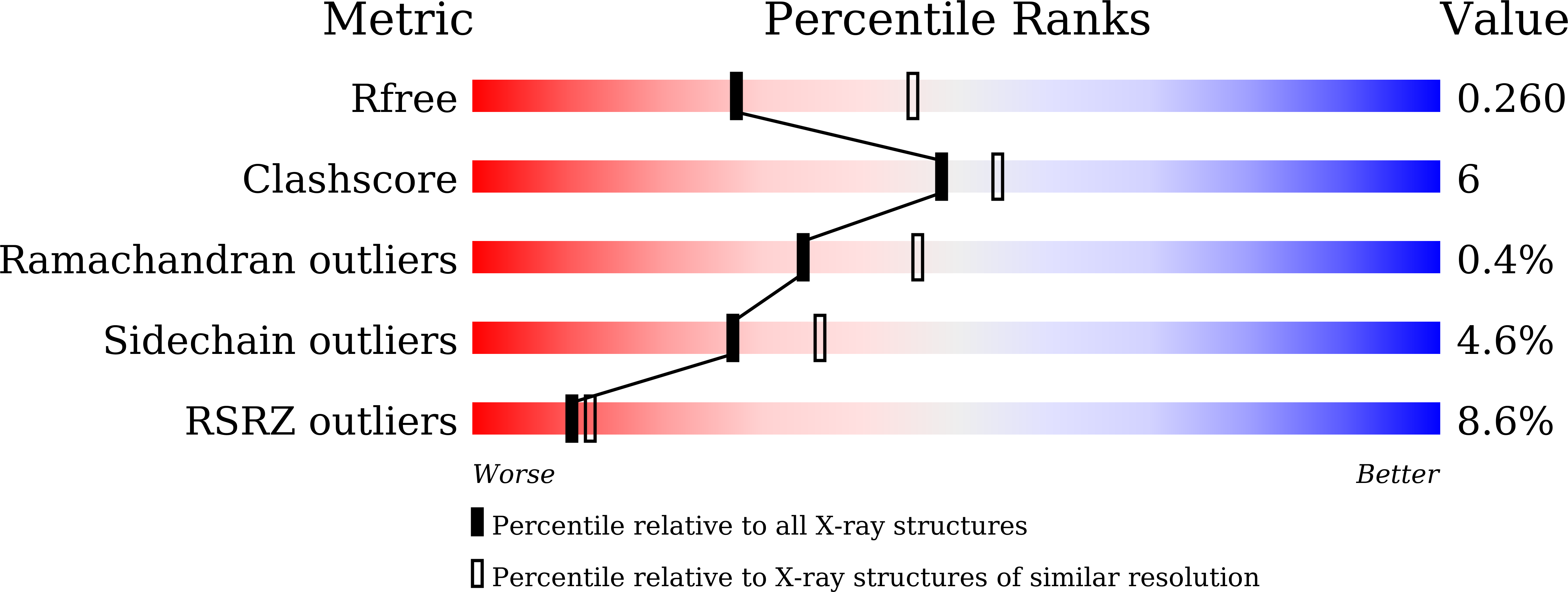
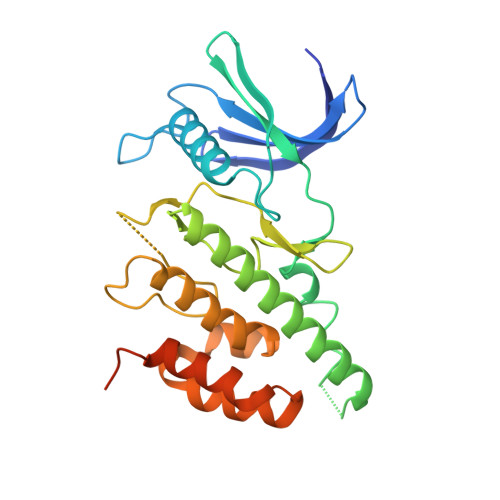
7WJ6, 7WJ7 - PubMed Abstract:
Class III lanthipeptide synthetases catalyze the formation of lanthionine/methyllanthionine and labionin crosslinks. We present here the 2.40 Å resolution structure of the kinase domain of a class III lanthipeptide synthetase CurKC from the biosynthesis of curvopeptin. A unique structural subunit for leader binding, named leader recognition domain (LRD), was identified. The LRD of CurKC is responsible for the recognition of the leader peptide and for mediating interactions between the lyase and kinase domains. LRDs are highly conserved among the kinase domains of class III and class IV lanthipeptide synthetases. The discovery of LRDs provides insight into the substrate recognition and domain organization in multidomain lanthipeptide synthetases.
Organizational Affiliation:
State Key Laboratory of Coordination Chemistry, Chemistry and Biomedicine Innovation Center of Nanjing University, Jiangsu Key Laboratory of Advanced Organic Materials, School of Chemistry and Chemical Engineering, Nanjing University, No. 163 Xianlin Ave, Nanjing, 210093, China.