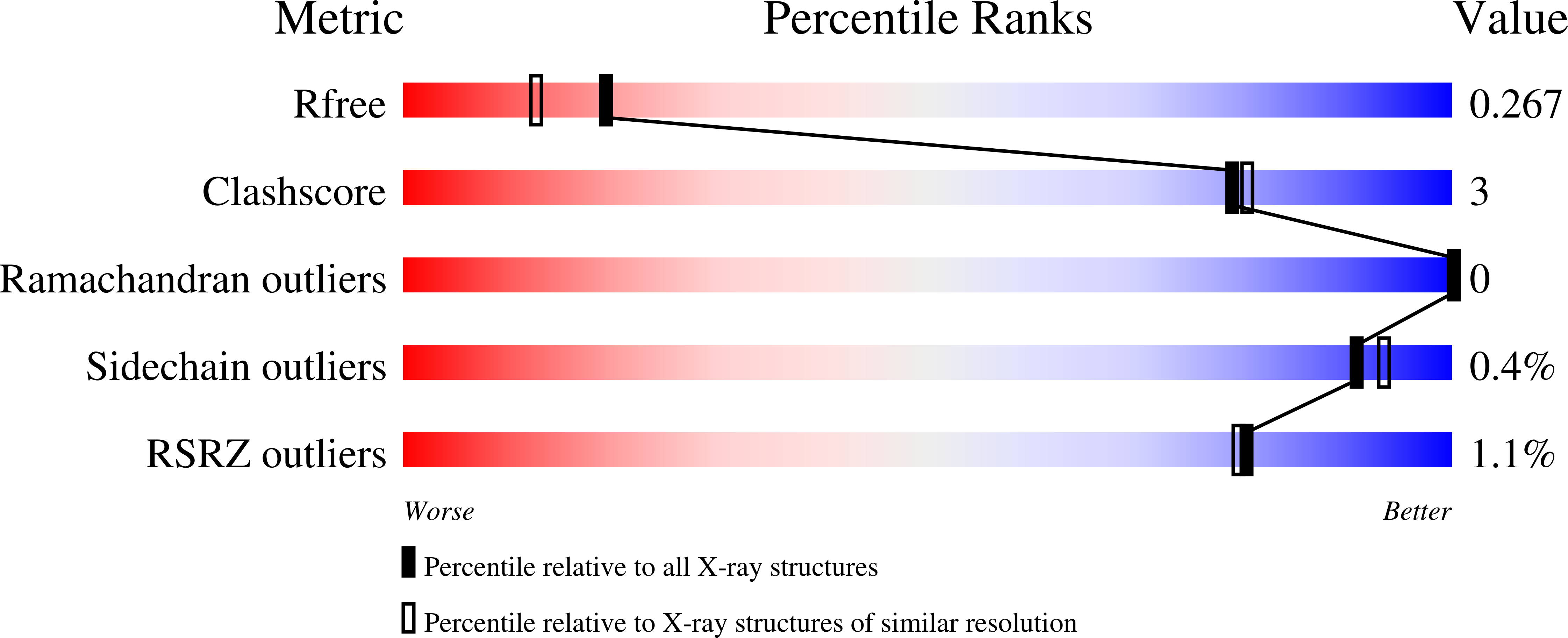
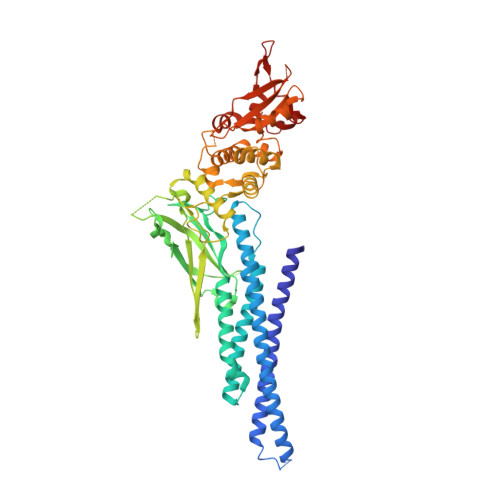
Poxviruses and paramyxoviruses use a conserved mechanism of STAT1 antagonism to inhibit interferon signaling.
Talbot-Cooper, C., Pantelejevs, T., Shannon, J.P., Cherry, C.R., Au, M.T., Hyvonen, M., Hickman, H.D., Smith, G.L.(2022) Cell Host Microbe 30: 357-372.e11
- PubMed: 35182467
- DOI: https://doi.org/10.1016/j.chom.2022.01.014
- Primary Citation of Related Structures:
7NUF - PubMed Abstract:
The induction of interferon (IFN)-stimulated genes by STATs is a critical host defense mechanism against virus infection. Here, we report that a highly expressed poxvirus protein, 018, inhibits IFN-induced signaling by binding to the SH2 domain of STAT1, thereby preventing the association of STAT1 with an activated IFN receptor. Despite encoding other inhibitors of IFN-induced signaling, a poxvirus mutant lacking 018 was attenuated in mice. The 2.0 Å crystal structure of the 018:STAT1 complex reveals a phosphotyrosine-independent mode of 018 binding to the SH2 domain of STAT1. Moreover, the STAT1-binding motif of 018 shows similarity to the STAT1-binding proteins from Nipah virus, which, similar to 018, block the association of STAT1 with an IFN receptor. Overall, these results uncover a conserved mechanism of STAT1 antagonism that is employed independently by distinct virus families.
Organizational Affiliation:
Department of Pathology, University of Cambridge, Tennis Court Road, Cambridge CB2 1QP, UK.