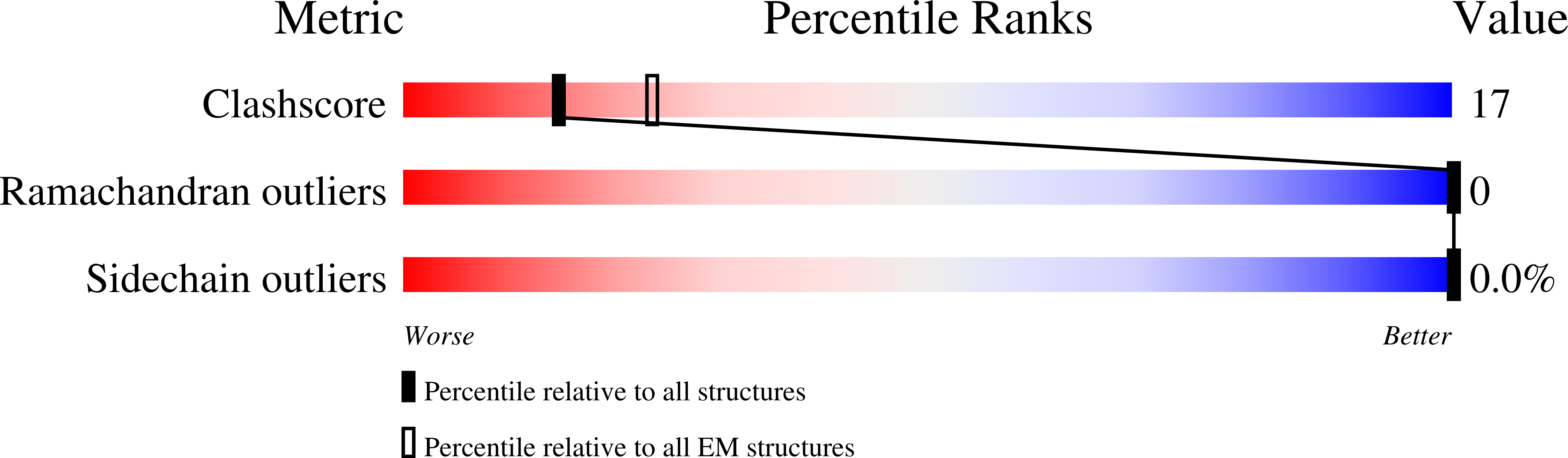Structural basis for inhibition of the AAA-ATPase Drg1 by diazaborine.
Prattes, M., Grishkovskaya, I., Hodirnau, V.V., Rossler, I., Klein, I., Hetzmannseder, C., Zisser, G., Gruber, C.C., Gruber, K., Haselbach, D., Bergler, H.(2021) Nat Commun 12: 3483-3483
- PubMed: 34108481
- DOI: https://doi.org/10.1038/s41467-021-23854-x
- Primary Citation of Related Structures:
7NKU - PubMed Abstract:
The hexameric AAA-ATPase Drg1 is a key factor in eukaryotic ribosome biogenesis and initiates cytoplasmic maturation of the large ribosomal subunit by releasing the shuttling maturation factor Rlp24. Drg1 monomers contain two AAA-domains (D1 and D2) that act in a concerted manner. Rlp24 release is inhibited by the drug diazaborine which blocks ATP hydrolysis in D2. The mode of inhibition was unknown. Here we show the first cryo-EM structure of Drg1 revealing the inhibitory mechanism. Diazaborine forms a covalent bond to the 2'-OH of the nucleotide in D2, explaining its specificity for this site. As a consequence, the D2 domain is locked in a rigid, inactive state, stalling the whole Drg1 hexamer. Resistance mechanisms identified include abolished drug binding and altered positioning of the nucleotide. Our results suggest nucleotide-modifying compounds as potential novel inhibitors for AAA-ATPases.
Organizational Affiliation:
Institute of Molecular Biosciences, University of Graz, Graz, Austria.