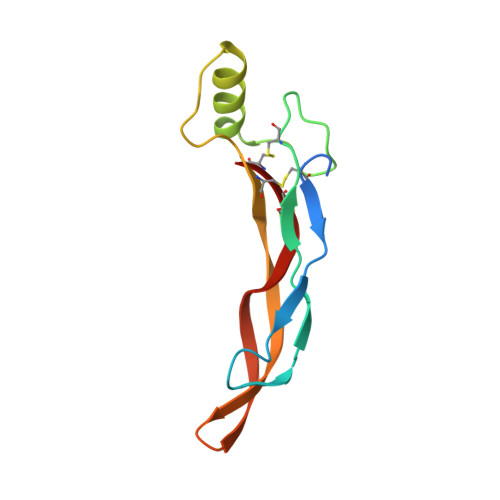
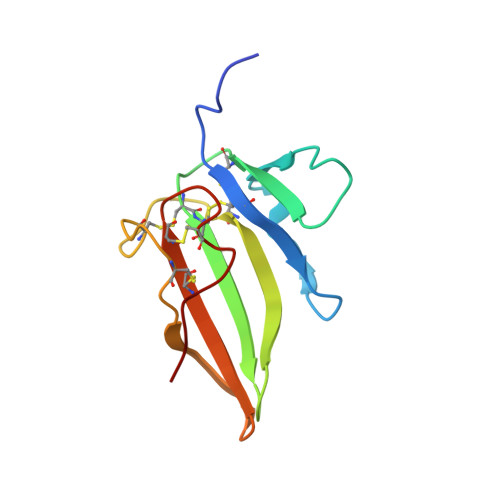
Structure of AMH bound to AMHR2 provides insight into a unique signaling pair in the TGF-beta family.
Hart, K.N., Stocker, W.A., Nagykery, N.G., Walton, K.L., Harrison, C.A., Donahoe, P.K., Pepin, D., Thompson, T.B.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34155118
- DOI: https://doi.org/10.1073/pnas.2104809118
- Primary Citation of Related Structures:
7L0J - PubMed Abstract:
Anti-Müllerian hormone (AMH), or Müllerian-inhibiting substance, is a protein hormone that promotes Müllerian duct regression during male fetal sexual differentiation and regulation of folliculogenesis in women. AMH is a member of the transforming growth factor beta (TGF-β) family, which has evolved to signal through its own dedicated type II receptor, AMH receptor type II (AMHR2). Structures of other TGF-β family members have revealed how ligands infer specificity for their cognate receptors; however, it is unknown how AMH binds AMHR2 at the molecular level. Therefore, in this study, we solved the X-ray crystal structure of AMH bound to the extracellular domain of AMHR2 to a resolution of 2.6Å. The structure reveals that while AMH binds AMHR2 in a similar location to Activin and BMP ligand binding to their type II receptors, differences in both AMH and AMHR2 account for a highly specific interaction. Furthermore, using an AMH responsive cell-based luciferase assay, we show that a conformation in finger 1 of AMHR2 and a salt bridge formed by K534 on AMH and D81/E84 of AMHR2 are key to the AMH/AMHR2 interaction. Overall, our study highlights how AMH engages AMHR2 using a modified paradigm of receptor binding facilitated by modifications to the three-finger toxin fold of AMHR2. Furthermore, understanding these elements contributing to the specificity of binding will help in the design of agonists or antagonists or the selection of antibody therapies.
Organizational Affiliation:
Department of Pharmacology and Systems Physiology, University of Cincinnati, Cincinnati, OH 45267.