Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Soldano, A., Yao, H., Punchi Hewage, A.N.D., Meraz, K., Annor-Gyamfi, J.K., Bunce, R.A., Battaile, K.P., Lovell, S., Rivera, M.(2021) ACS Infect Dis 7: 123-140
- PubMed: 33269912
- DOI: https://doi.org/10.1021/acsinfecdis.0c00669
- Primary Citation of Related Structures:
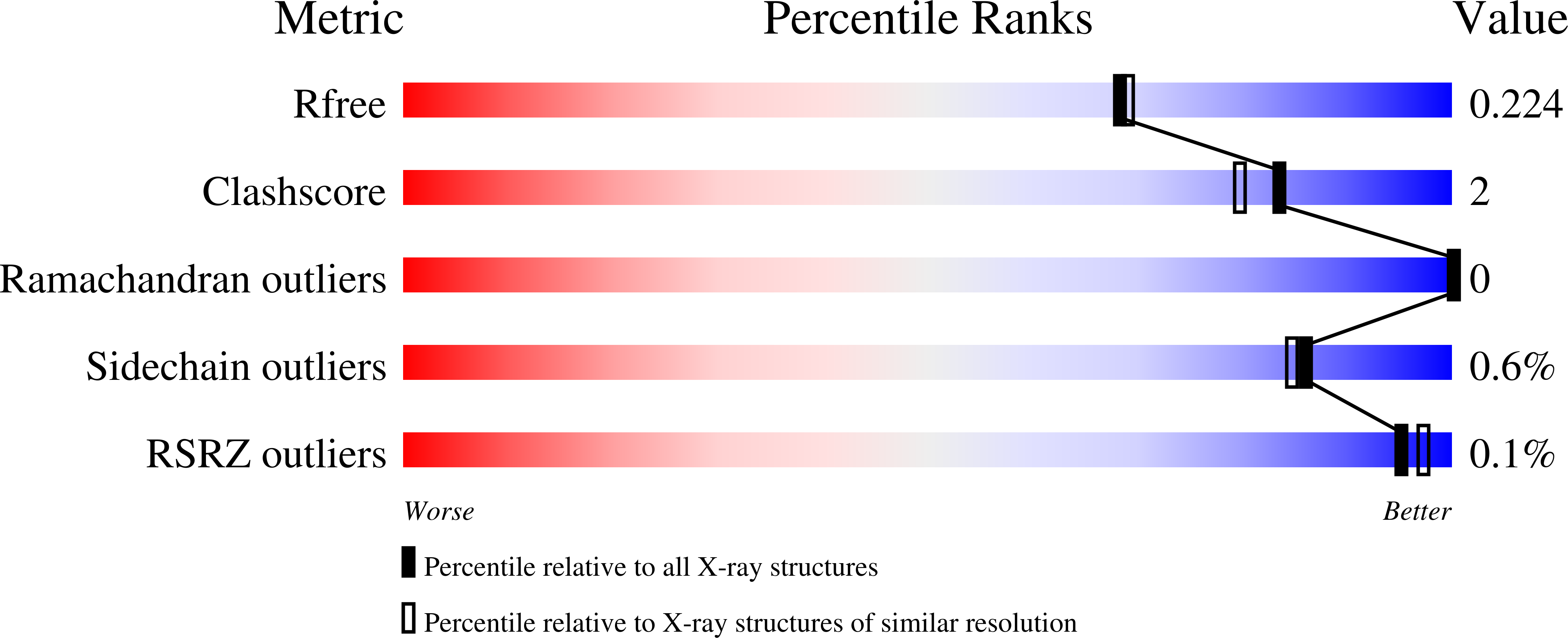
7K5E, 7K5F, 7K5G, 7K5H - PubMed Abstract:
Bacteria depend on a well-regulated iron homeostasis to survive adverse environments. A key component of the iron homeostasis machinery is the compartmentalization of Fe 3+ in bacterioferritin and its subsequent mobilization as Fe 2+ to satisfy metabolic requirements. In Pseudomonas aeruginosa Fe 3+ is compartmentalized in bacterioferritin (BfrB), and its mobilization to the cytosol requires binding of a ferredoxin (Bfd) to reduce the stored Fe 3+ and release the soluble Fe 2+ . Blocking the BfrB-Bfd complex in P. aeruginosa by deletion of the bfd gene triggers an irreversible accumulation of Fe 3+ in BfrB, concomitant cytosolic iron deficiency and significant impairment of biofilm development. Herein we report that small molecules developed to bind BfrB at the Bfd binding site block the BfrB-Bfd complex, inhibit the mobilization of iron from BfrB in P. aeruginosa cells, elicit a bacteriostatic effect on planktonic cells, and are bactericidal to cells embedded in mature biofilms.
Organizational Affiliation:
Department of Chemistry, Louisiana State University, 232 Choppin Hall, Baton Rouge, Louisiana 70803, United States.