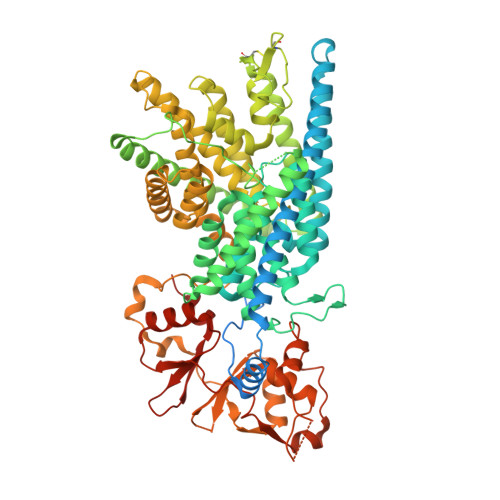
Cryo-EM structure of the lysosomal chloride-proton exchanger CLC-7 in complex with OSTM1.
Schrecker, M., Korobenko, J., Hite, R.K.(2020) Elife 9
- PubMed: 32749217
- DOI: https://doi.org/10.7554/eLife.59555
- Primary Citation of Related Structures:
7JM6, 7JM7 - PubMed Abstract:
The chloride-proton exchanger CLC-7 plays critical roles in lysosomal homeostasis and bone regeneration and its mutation can lead to osteopetrosis, lysosomal storage disease and neurological disorders. In lysosomes and the ruffled border of osteoclasts, CLC-7 requires a β-subunit, OSTM1, for stability and activity. Here, we present electron cryomicroscopy structures of CLC-7 in occluded states by itself and in complex with OSTM1, determined at resolutions up to 2.8 Å. In the complex, the luminal surface of CLC-7 is entirely covered by a dimer of the heavily glycosylated and disulfide-bonded OSTM1, which serves to protect CLC-7 from the degradative environment of the lysosomal lumen. OSTM1 binding does not induce large-scale rearrangements of CLC-7, but does have minor effects on the conformation of the ion-conduction pathway, potentially contributing to its regulatory role. These studies provide insights into the role of OSTM1 and serve as a foundation for understanding the mechanisms of CLC-7 regulation.
Organizational Affiliation:
Structural Biology Program, Memorial Sloan Kettering Cancer Center, New York, United States.