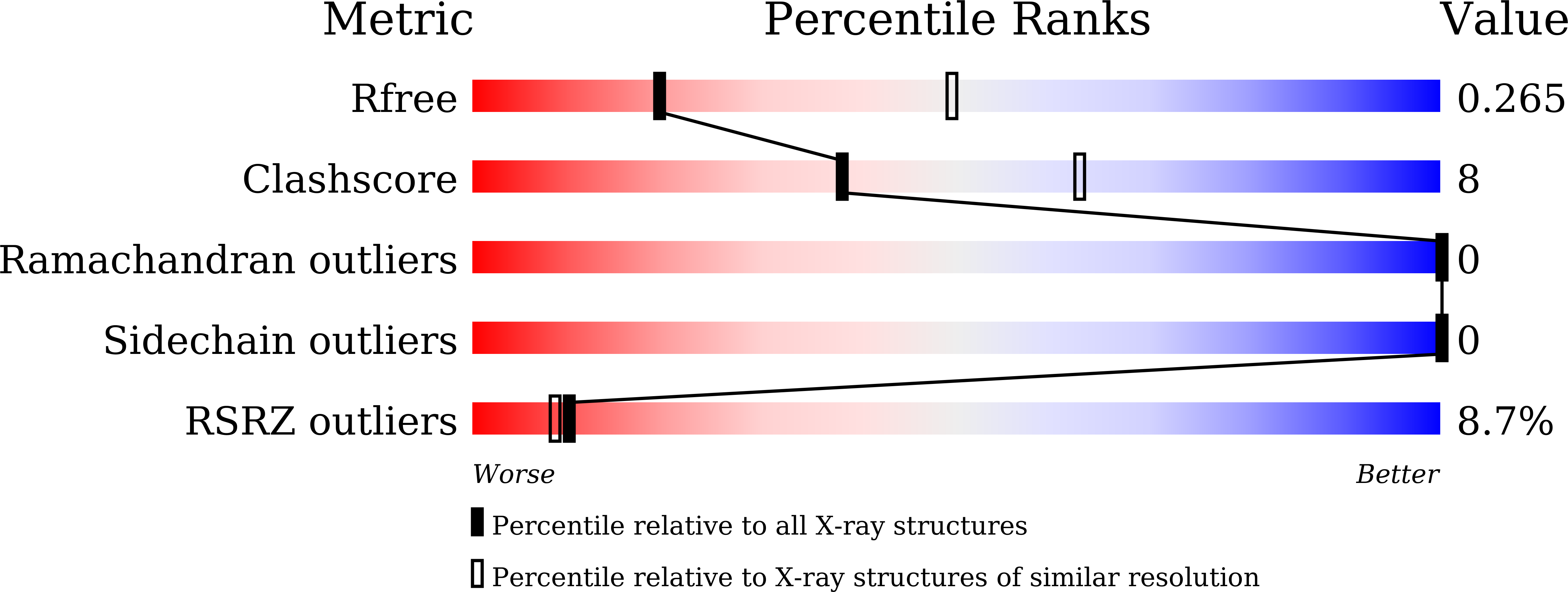
Molecular mechanism of agonism and inverse agonism in ghrelin receptor.
Qin, J., Cai, Y., Xu, Z., Ming, Q., Ji, S.Y., Wu, C., Zhang, H., Mao, C., Shen, D.D., Hirata, K., Ma, Y., Yan, W., Zhang, Y., Shao, Z.(2022) Nat Commun 13: 300-300
- PubMed: 35027551
- DOI: https://doi.org/10.1038/s41467-022-27975-9
- Primary Citation of Related Structures:
7F83, 7W2Z - PubMed Abstract:
Much effort has been invested in the investigation of the structural basis of G protein-coupled receptors (GPCRs) activation. Inverse agonists, which can inhibit GPCRs with constitutive activity, are considered useful therapeutic agents, but the molecular mechanism of such ligands remains insufficiently understood. Here, we report a crystal structure of the ghrelin receptor bound to the inverse agonist PF-05190457 and a cryo-electron microscopy structure of the active ghrelin receptor-Go complex bound to the endogenous agonist ghrelin. Our structures reveal a distinct binding mode of the inverse agonist PF-05190457 in the ghrelin receptor, different from the binding mode of agonists and neutral antagonists. Combining the structural comparisons and cellular function assays, we find that a polar network and a notable hydrophobic cluster are required for receptor activation and constitutive activity. Together, our study provides insights into the detailed mechanism of ghrelin receptor binding to agonists and inverse agonists, and paves the way to design specific ligands targeting ghrelin receptors.
Organizational Affiliation:
Department of Biophysics and Department of Pathology of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, 310058, Hangzhou, China.