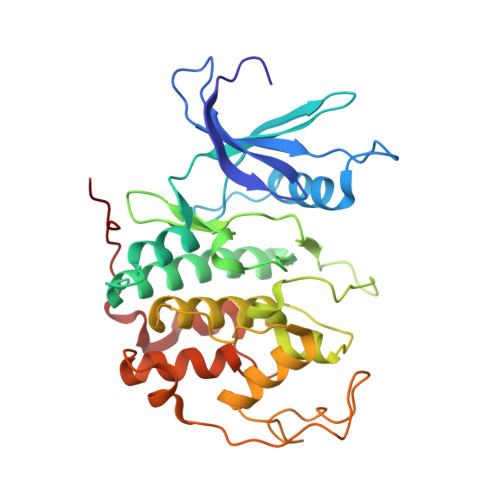
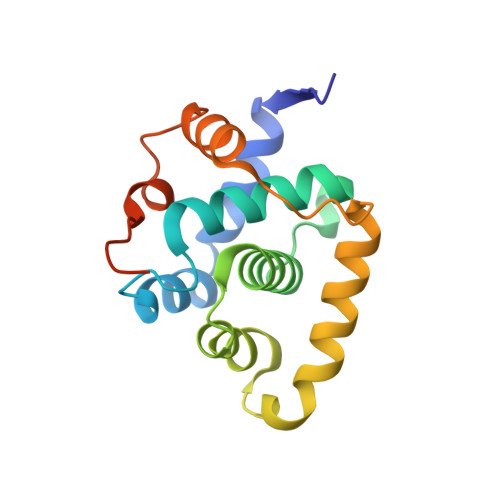
The SUN1-SPDYA interaction plays an essential role in meiosis prophase I.
Chen, Y., Wang, Y., Chen, J., Zuo, W., Fan, Y., Huang, S., Liu, Y., Chen, G., Li, Q., Li, J., Wu, J., Bian, Q., Huang, C., Lei, M.(2021) Nat Commun 12: 3176-3176
- PubMed: 34039995
- DOI: https://doi.org/10.1038/s41467-021-23550-w
- Primary Citation of Related Structures:
7E34 - PubMed Abstract:
Chromosomes pair and synapse with their homologous partners to segregate correctly at the first meiotic division. Association of telomeres with the LINC (Linker of Nucleoskeleton and Cytoskeleton) complex composed of SUN1 and KASH5 enables telomere-led chromosome movements and telomere bouquet formation, facilitating precise pairwise alignment of homologs. Here, we identify a direct interaction between SUN1 and Speedy A (SPDYA) and determine the crystal structure of human SUN1-SPDYA-CDK2 ternary complex. Analysis of meiosis prophase I process in SPDYA-binding-deficient SUN1 mutant mice reveals that the SUN1-SPDYA interaction is required for the telomere-LINC complex connection and the assembly of a ring-shaped telomere supramolecular architecture at the nuclear envelope, which is critical for efficient homologous pairing and synapsis. Overall, our results provide structural insights into meiotic telomere structure that is essential for meiotic prophase I progression.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, China.