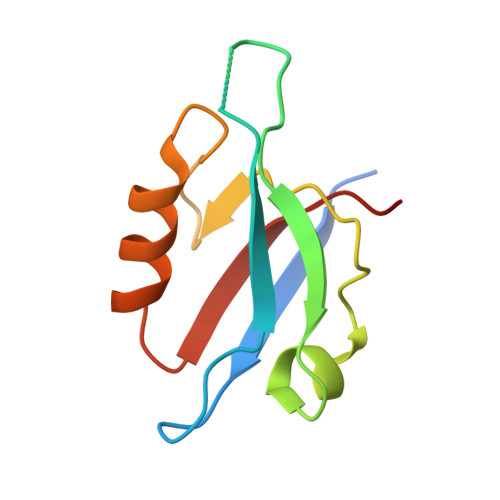
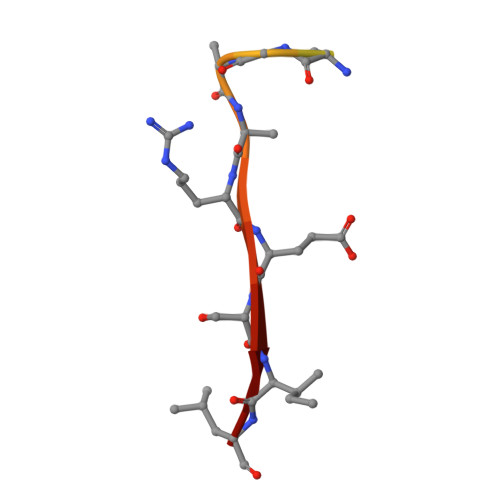
Crystal structure of the PDZ4 domain of MAGI2 in complex with PBM of ARMS reveals a canonical PDZ recognition mode.
Zhang, Y., Zhong, Z., Ye, J., Wang, C.(2021) Neurochem Int 149: 105152-105152
- PubMed: 34371146
- DOI: https://doi.org/10.1016/j.neuint.2021.105152
- Primary Citation of Related Structures:
7D6F - PubMed Abstract:
Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (MAGI2) is a neuronal scaffold protein that plays critical roles at synaptic junctions by assembling neurotransmitter receptors and cell adhesion proteins through its multiple protein-protein interaction domains, including six PDZ domains, two phosphoserine-phosphothreonine binding WW domains, and a guanylate kinase GK domain. Previous studies showed that MAGI2 participates in formation of tetrameric complexes with PDZ-GEF1, TrkA receptor, and ankyrin repeat-rich membrane spanning (ARMS) protein at late endosomes and is crucial for neurite outgrowth. However, the molecular mechanism governing the assembly of these complexes remains unknown. Here, we characterize the direct interaction between MAGI2 and ARMS through multiple biochemical assays. Moreover, our solved crystal structure of the truncated PDZ4/PBM (PDZ binding motifs) complex of MAGI2 and ARMS proteins (MAGI2-PDZ4/ARMS-PBM) reveals that the binding interface lies between the αB/βB groove from the PDZ4 of MAGI2 and the C-terminal PBM from ARMS. The structure reveals high similarity to others in this protein family where canonical PDZ/PBM interactions are observed. However, the conserved "GLGF" motif in the PSD-95-PDZ3 changes to "GFGF" in the MAGI2-PDZ4/ARMS-PBM complex. We further validated our crystal structure through serial mutagenesis assays. Taken together, our study provides the biochemical details and binding mechanisms that underpin the stabilization of the MAGI2-PDZ4/ARMS-PBM complex, thereby offering a biochemical and structural basis for further understanding of the functional roles of MAGI2, ARMS, PDZ-GEF1, and TrkA in forming the tetrameric receptor complex in neuronal signaling.
Organizational Affiliation:
Department of Neurology, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, MOE Key Laboratory for Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, University of Science and Technology of China, 230001, Hefei, China.