Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate).
Suzuki, K., Koitabashi, M.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cutinase | 195 | Paraphoma sp. B47-9 | Mutation(s): 0 EC: 3.1.1.74 | 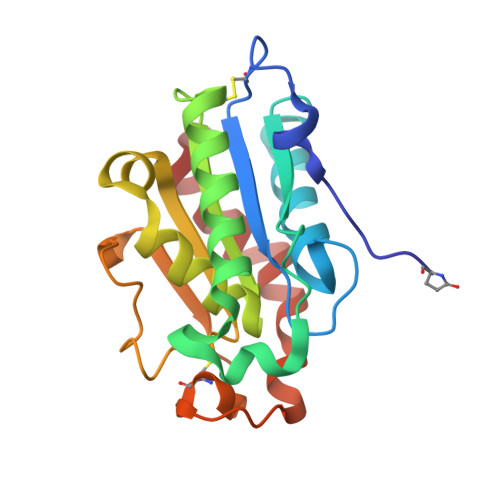 | |
UniProt | |||||
Find proteins for A0A060N399 (Paraphoma sp. B47-9) Explore A0A060N399 Go to UniProtKB: A0A060N399 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A060N399 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CAD (Subject of Investigation/LOI) Query on CAD | C [auth A], D [auth A], G [auth B] | CACODYLIC ACID C2 H7 As O2 OGGXGZAMXPVRFZ-UHFFFAOYSA-N |  | ||
| NA Query on NA | E [auth A], F [auth A], H [auth B], I [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| PCA Query on PCA | A, B | L-PEPTIDE LINKING | C5 H7 N O3 |  | GLN |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 34.886 | α = 103.78 |
| b = 43.517 | β = 92.42 |
| c = 51.724 | γ = 101.49 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| Aimless | data scaling |
| SHELX | phasing |
| PDB_EXTRACT | data extraction |
| xia2 | data reduction |