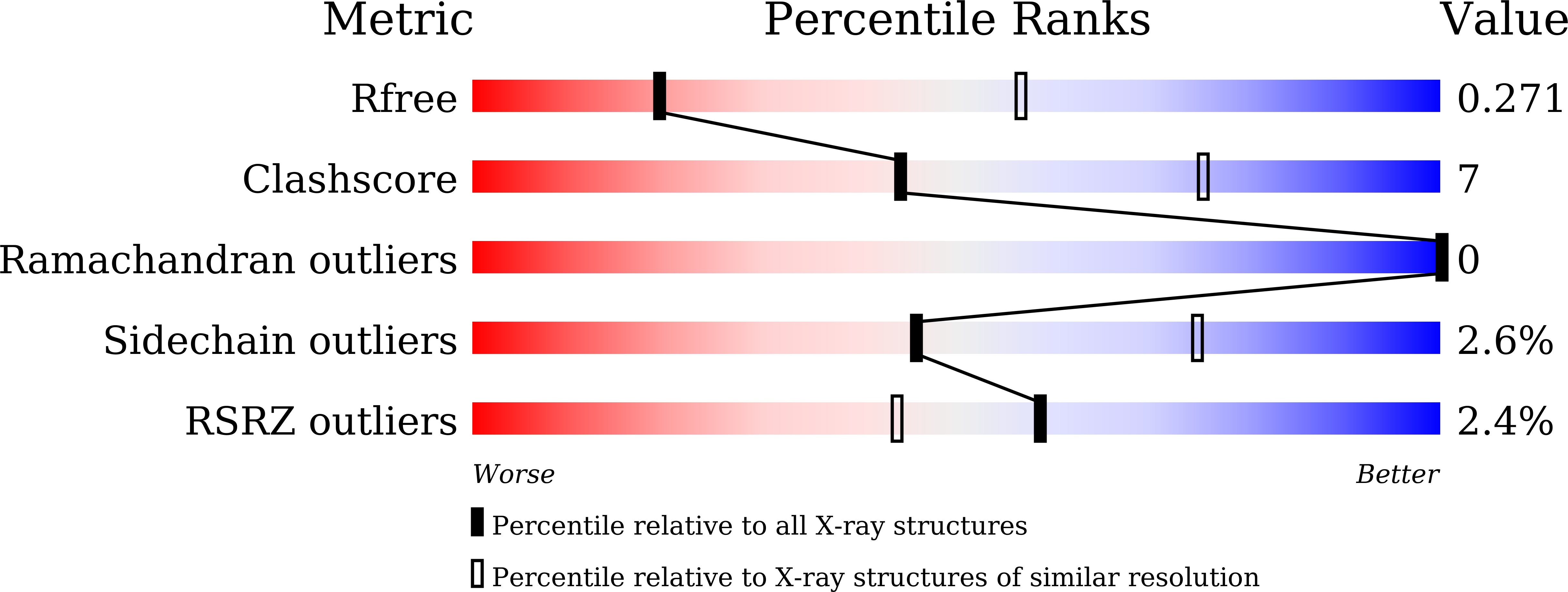
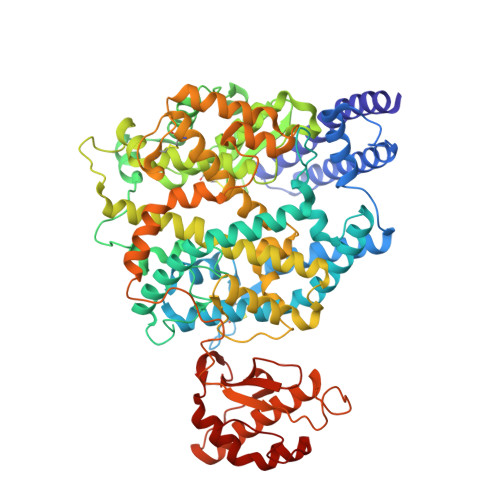
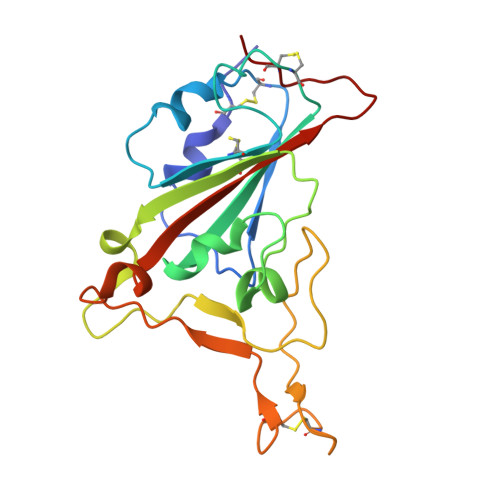
Cross-species recognition of SARS-CoV-2 to bat ACE2.
Liu, K., Tan, S., Niu, S., Wang, J., Wu, L., Sun, H., Zhang, Y., Pan, X., Qu, X., Du, P., Meng, Y., Jia, Y., Chen, Q., Deng, C., Yan, J., Wang, H.W., Wang, Q., Qi, J., Gao, G.F.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33335073
- DOI: https://doi.org/10.1073/pnas.2020216118
- Primary Citation of Related Structures:
7C8J, 7C8K - PubMed Abstract:
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has emerged as a major threat to global health. Although varied SARS-CoV-2-related coronaviruses have been isolated from bats and SARS-CoV-2 may infect bat, the structural basis for SARS-CoV-2 to utilize the human receptor counterpart bat angiotensin-converting enzyme 2 (bACE2) for virus infection remains less understood. Here, we report that the SARS-CoV-2 spike protein receptor binding domain (RBD) could bind to bACE2 from Rhinolophus macrotis (bACE2-Rm) with substantially lower affinity compared with that to the human ACE2 (hACE2), and its infectivity to host cells expressing bACE2-Rm was confirmed with pseudotyped SARS-CoV-2 virus and SARS-CoV-2 wild virus. The structure of the SARS-CoV-2 RBD with the bACE2-Rm complex was determined, revealing a binding mode similar to that of hACE2. The analysis of binding details between SARS-CoV-2 RBD and bACE2-Rm revealed that the interacting network involving Y41 and E42 of bACE2-Rm showed substantial differences with that to hACE2. Bats have extensive species diversity and the residues for RBD binding in bACE2 receptor varied substantially among different bat species. Notably, the Y41H mutant, which exists in many bats, attenuates the binding capacity of bACE2-Rm, indicating the central roles of Y41 in the interaction network. These findings would benefit our understanding of the potential infection of SARS-CoV-2 in varied species of bats.
Organizational Affiliation:
Chinese Academy of Sciences Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, 100101 Beijing, China.