Structure and Function Insight of the alpha-Glucosidase QsGH13 From Qipengyuania seohaensis sp. SW-135.
Zhai, X., Wu, K., Ji, R., Zhao, Y., Lu, J., Yu, Z., Xu, X., Huang, J.(2022) Front Microbiol 13: 849585-849585
- PubMed: 35308395
- DOI: https://doi.org/10.3389/fmicb.2022.849585
- Primary Citation of Related Structures:
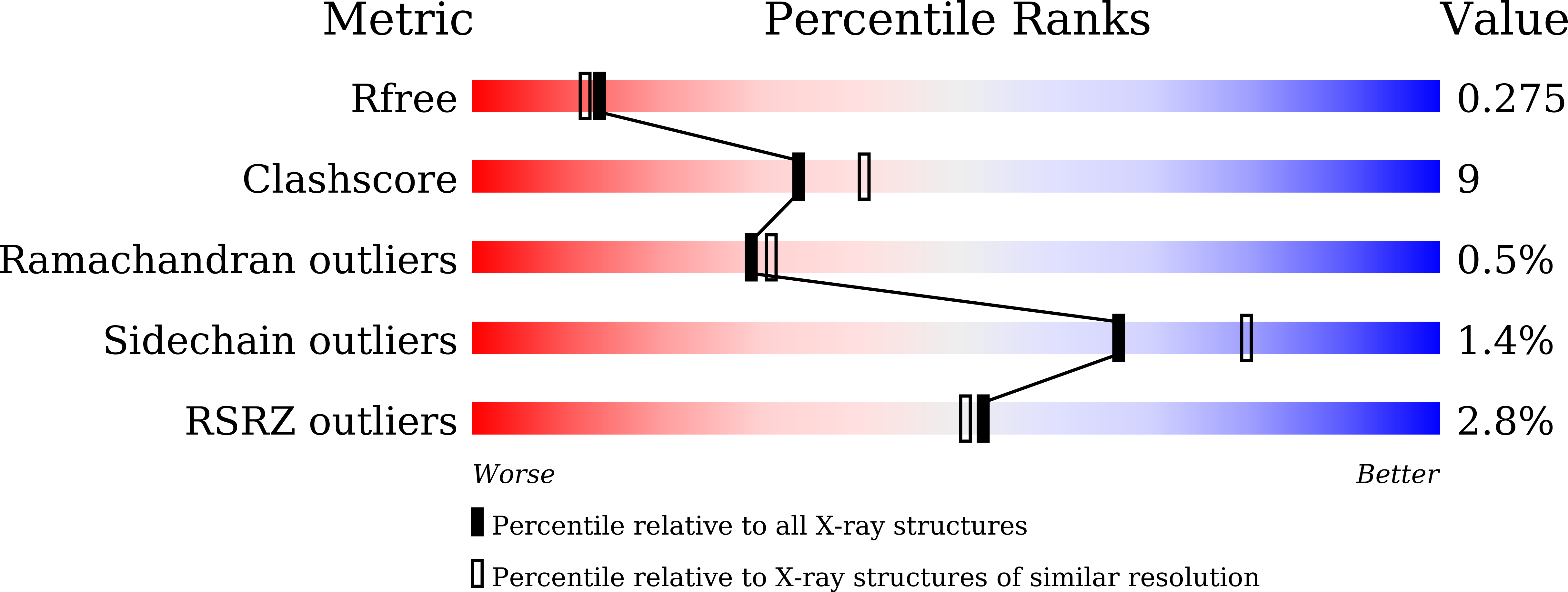
7VOH - PubMed Abstract:
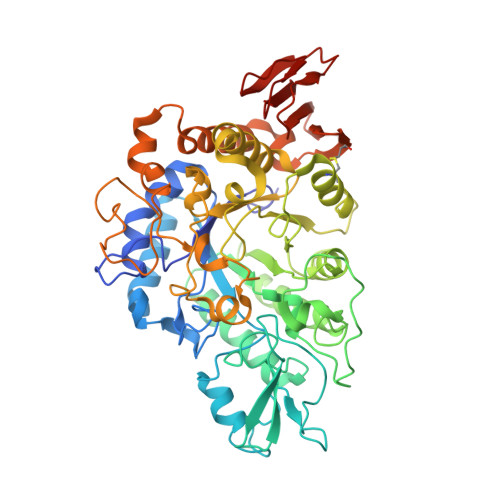
The α-glucosidases play indispensable roles in the metabolic mechanism of organism, prevention, and treatment of the disease, and sugar hydrolysis, and are widely used in chemical synthesis, clinical diagnosis, and other fields. However, improving their catalytic efficiency and production to meet commercial demand remains a huge challenge. Here we detected a novel GH13 family α-glucosidase, QsGH13, from the deep-sea bacterium Qipengyuania seohaensis sp. SW-135. QsGH13 is highly substrate specific and only hydrolyzes sugars containing alpha-1,4 glucoside bonds. For example, its enzymatic activity for p -nitrophenyl-α-D-glucopyranoside was 25.41 U/mg, and the K m value was 0.2952 ± 0.0322 mM. The biochemical results showed that the optimum temperature of QsGH13 is 45°C, the optimum pH is 10.0, and it has excellent biological characteristics such as alkali resistance and salt resistance. The crystal structure of QsGH13 was resolved with a resolution of 2.2 Å, where QsGH13 is composed of a typical TIM barrel catalytic domain A, a loop-rich domain B, and a conserved domain C. QsGH13 crystal belonged to the monoclinic space group P2 1 2 1 2 1 , with unit-cell parameters a = 58.816 Å, b = 129.920 Å, c = 161.307 Å, α = γ = β = 90°, which contains two monomers per asymmetric unit. The β → α loop 4 of QsGH13 was located above catalytic pocket. Typical catalytic triad residues Glu202, Asp266, and Glu329 were found in QsGH13. The biochemical properties and structural analysis of QsGH13 have greatly improved our understanding of the catalytic mechanism of GH13 family. This study provides new ideas to broaden the application of α-glucosidase in alcohol fermentation, glycolysis, and other industries.
Organizational Affiliation:
Department of Parasitology, School of Basic Medical Science, Central South University, Changsha, China.