Rationally designed immunogens enable immune focusing following SARS-CoV-2 spike imprinting.
Hauser, B.M., Sangesland, M., St Denis, K.J., Lam, E.C., Case, J.B., Windsor, I.W., Feldman, J., Caradonna, T.M., Kannegieter, T., Diamond, M.S., Balazs, A.B., Lingwood, D., Schmidt, A.G.(2022) Cell Rep 38: 110561-110561
- PubMed: 35303475
- DOI: https://doi.org/10.1016/j.celrep.2022.110561
- Primary Citation of Related Structures:
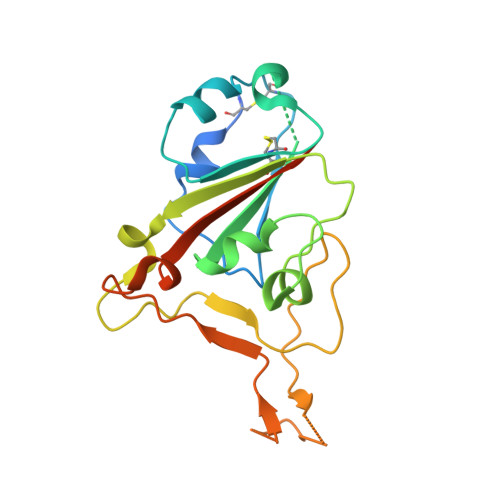
7TE1 - PubMed Abstract:
Eliciting antibodies to surface-exposed viral glycoproteins can generate protective responses that control and prevent future infections. Targeting conserved sites may reduce the likelihood of viral escape and limit the spread of related viruses with pandemic potential. Here we leverage rational immunogen design to focus humoral responses on conserved epitopes. Using glycan engineering and epitope scaffolding in boosting immunogens, we focus murine serum antibody responses to conserved receptor binding motif (RBM) and receptor binding domain (RBD) epitopes following severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike imprinting. Although all engineered immunogens elicit a robust SARS-CoV-2-neutralizing serum response, RBM-focusing immunogens exhibit increased potency against related sarbecoviruses, SARS-CoV, WIV1-CoV, RaTG13-CoV, and SHC014-CoV; structural characterization of representative antibodies defines a conserved epitope. RBM-focused sera confer protection against SARS-CoV-2 challenge. Thus, RBM focusing is a promising strategy to elicit breadth across emerging sarbecoviruses without compromising SARS-CoV-2 protection. These engineering strategies are adaptable to other viral glycoproteins for targeting conserved epitopes.
Organizational Affiliation:
Ragon Institute of MGH, MIT, and Harvard, Cambridge, MA 02139, USA.