Biotechnological potential and initial characterization of two novel sesquiterpene synthases from Basidiomycota Coniophora puteana for heterologous production of delta-cadinol.
Ringel, M., Dimos, N., Himpich, S., Haack, M., Huber, C., Eisenreich, W., Schenk, G., Loll, B., Bruck, T.(2022) Microb Cell Fact 21: 64-64
- PubMed: 35440053
- DOI: https://doi.org/10.1186/s12934-022-01791-8
- Primary Citation of Related Structures:
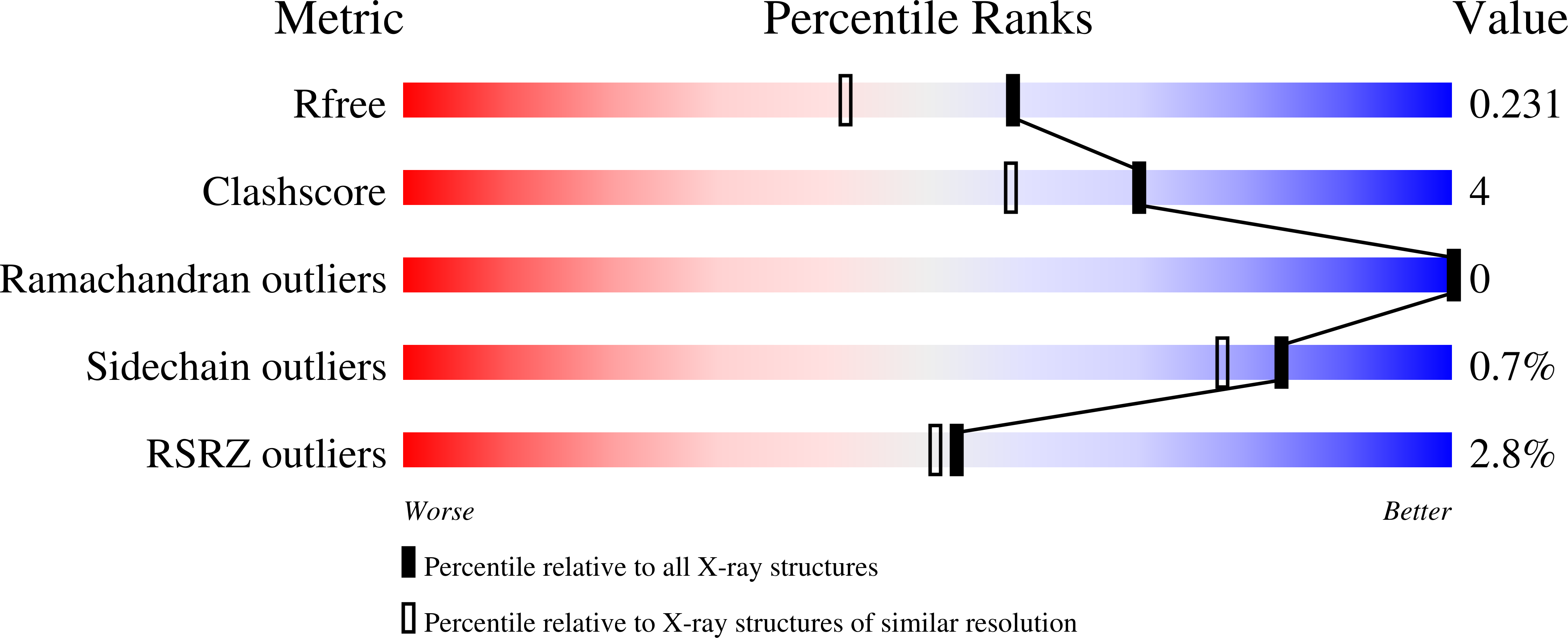
7OFL - PubMed Abstract:
Terpene synthases are versatile catalysts in all domains of life, catalyzing the formation of an enormous variety of different terpenoid secondary metabolites. Due to their diverse bioactive properties, terpenoids are of great interest as innovative ingredients in pharmaceutical and cosmetic applications. Recent advances in genome sequencing have led to the discovery of numerous terpene synthases, in particular in Basidiomycota like the wood rotting fungus Coniophora puteana, which further enhances the scope for the manufacture of terpenes for industrial purposes. In this study we describe the identification of two novel (+)-δ-cadinol synthases from C. puteana, Copu5 and Copu9. The sesquiterpene (+)-δ-cadinol was previously shown to exhibit cytotoxic activity therefore having an application as possible, new, and sustainably sourced anti-tumor agent. In an Escherichia coli strain, optimized for sesquiterpene production, titers of 225 mg l -1 and 395 mg l -1 , respectively, could be achieved. Remarkably, both enzymes share the same product profile thereby representing the first two terpene synthases from Basidiomycota with identical product profiles. We solved the crystal structure of Copu9 in its closed conformation, for the first time providing molecular details of sesquiterpene synthase from Basidiomycota. Based on the Copu9 structure, we conducted structure-based mutagenesis of amino acid residues lining the active site, thereby altering the product profile. Interestingly, the mutagenesis study also revealed that despite the conserved product profiles of Copu5 and Copu9 different conformational changes may accompany the catalytic cycle of the two enzymes. This observation suggests that the involvement of tertiary structure elements in the reaction mechanism(s) employed by terpene synthases may be more complex than commonly expected. The presented product selectivity and titers of Copu5 and Copu9 may pave the way towards a sustainable, biotechnological production of the potentially new bioactive (+)-δ-cadinol. Furthermore, Copu5 and Copu9 may serve as model systems for further mechanistic studies of terpenoid catalysis.
Organizational Affiliation:
Werner Siemens Chair of Synthetic Biotechnology, Department of Chemistry, Technical University of Munich, Lichtenbergstr. 4, 85748, Garching, Germany.